Abstract
Arsenic groundwater contamination threatens the health of millions of people worldwide, particularly in South and Southeast Asia. In most cases, the release of arsenic from sediment was caused by microbial reductive dissolution of arsenic-bearing iron(III) minerals with organic carbon being used as microbial electron donor. Although in many arsenic-contaminated aquifers high concentrations of methane were observed, its role in arsenic mobilization is unknown. Here, using microcosms experiments and hydrogeochemical and microbial community analyses, we demonstrate that methane functions as electron donor for methanotrophs, triggering the reductive dissolution of arsenic-bearing iron(III) minerals, increasing the abundance of genes related to methane oxidation, and ultimately mobilizing arsenic into the water. Our findings provide evidence for a methane-mediated mechanism for arsenic mobilization that is distinct from previously described pathways. Taking this together with the common presence of methane in arsenic-contaminated aquifers, we suggest that this methane-driven arsenic mobilization may contribute to arsenic contamination of groundwater on a global scale.
Similar content being viewed by others
Introduction
Exposure to groundwater contaminated with arsenic (As) is a worldwide problem. Arsenical skin lesions are the hallmark of chronic arsenic poisoning appearing within a few years of exposure often leading to skin cancer1. These symptoms are prevalent in populations living along river floodplains of South and Southeast Asia that rely on shallow groundwater wells for drinking water and irrigation2,3. Arsenic enrichment in shallow groundwater4 has resulted in the so-called “worst mass poisoning of human population in history”5. Generally, Fe(III) (oxyhydr)oxides are common constituents of As-bearing aquifer sediments6. Various mechanisms of As release to the groundwater have been suggested including abiotic dissolution and transformation of As-containing Fe minerals including oxidation of arsenic-bearing sulfides and changes in sorption capacity of Fe(III) (oxyhydr)oxide minerals as well as As desorption due to pH variations or by competition with phosphate7. Yet, the most commonly accepted mechanism is microbial reductive dissolution of As-bearing Fe(III) (oxyhydr)oxides8,9,10. Fe(III)-reducing bacteria such as Geobacter, however, require bioavailable carbon (C) for their activity11,12. Previous studies focused on the identity and source of organic compounds used for microbial As mobilization13,14,15,16; however, the role of methane (CH4) remained unexplored. Here we investigated whether CH4, which is abundant in these aquifers, can also be a potential energy source and drive microbially mediated Fe(III)-mineral reduction leading to As mobilization.
Results and discussion
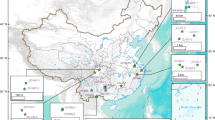
Van Phuc, a village 15 km SE from Hanoi, is known for severe As groundwater contamination (Fig. 1a), sometimes 60-fold higher than the World Health Organization (WHO) drinking water limit of 10 µg/L17. Arsenic-bearing Fe(III) minerals18,19 and groundwater containing high As, Fe, and CH4, low SO42−, and no NO3− were reported (Fig. 1b)20. In situ gas measurements demonstrated that the Total dissolved gas pressure (TDGP) correlates with As concentrations and CH4 accounts for the majority of detected gases in the groundwater (Fig. 1c). This makes Van Phuc an ideal location to study Fe(III)-dependent anaerobic methane oxidation and its relevance for As mobilization. Analysis of the in situ microbial community composition by 16S rRNA gene amplicon sequencing in aquifer sediments and groundwater (Fig. 2) showed that fermenting, methanogenic, and methanotrophic microorganisms dominated sediments and groundwater, suggesting that CH4 cycling occurs in this aquifer. Moreover, in some of the groundwater wells we found high abundances (up to 4%) of sequences affiliated to Candidatus Methanoperedens, a known Fe(III)-reducing CH4 oxidizer21,22.
a Distribution of monitoring wells and b concentrations of CH4, Fe, and As in groundwater. Wells with high CH4 are also characterized by high dissolved Fe and As (in bold red). c Total dissolved gas pressure of which the majority accounts for CH4 correlates with As in groundwater. Groundwater data from Stopelli et al. (2020) (ref. 20). Satellite image: Google, Maxar Technologies.
a Sedimentary microbial community from different depths across the redox transition zone. The presence of fermenters suggests organic carbon degradation across the aquifer and microorganisms likely involved in CH4 cycling. b Groundwater microbial community from monitoring wells. Relative abundance of microorganisms likely involved in CH4 cycling compared to microorganisms potentially involved in Fe cycling indicates the importance of CH4 as an electron donor in this aquifer.
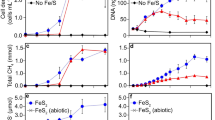
Using As- and Fe-bearing sediments (Supplementary Fig. 1) (~40 m depth), we investigated whether CH4 can induce microbially driven Fe(III) mineral reduction and As mobilization. Microbially active sediments were amended with artificial groundwater and field-relevant CH4 concentrations (~45 mg CH4/L) and compared to abiotic controls (including CH4) and biotic controls (without CH4). Fe redox speciation in water and sediments (quantified using the spectrophotometric Ferrozine assay), dissolved and solid-phase arsenic (analyzed by inductively coupled plasma mass spectrometry (ICP-MS) and by synchrotron-based X-ray absorption near-edge structure (XANES) spectroscopy), as well as major ions (determined by ICP-MS) were monitored over time (for more details see Supplementary Figs. 1–4). Due to the high CH4 and CO2 concentrations (simulating in situ field conditions), we were unable to monitor changes in their concentrations (Supplementary Note 1). We characterized the microbial community and identified microorganisms mediating CH4 oxidation and Fe(III) reduction after 125 days and 220 days of incubation. These time points were chosen based on their obvious relevance regarding the Fe(III) reduction process (Fig. 3). The results showed that CH4 triggers Fe(III)-mineral reduction, induces changes in the microbial community composition, increases the abundance of methane-cycle mcrA and pmoA genes, and ultimately releases As into the water. Our study therefore for the first time provides experimental evidence for As mobilization via CH4 oxidation coupled to Fe(III) minerals reduction.
a Sedimentary Fe(II), b dissolved Fe2+, and c dissolved As over 220 days of incubation of As-bearing sediments supplied with CH4 compared to biotic and abiotic setups. Error bars represent standard deviation from six microcosms until 125 days and three microcosms from days 125 to 220. Each microcosm was measured in triplicate. Please note that in panel b the red symbols cover the black symbols (all concentrations are at a value of 0).
Specifically, we found that in the biotic microcosms with CH4, up to 0.5 mg Fe/g (max. 6% of the total sedimentary Fe) was reduced to Fe(II) until day 120 (Fig. 3a and Supplementary Fig. 2), but no Fe2+ was released into the water (Fig. 3b). Despite the low extent of Fe(III) reduction, As mobilization was observed, reaching nearly 1.5 µg/L (Fig. 3c), ca. 2% of total sedimentary As. This initial release of As suggests that this As was bound to the easily bioavailable fraction of Fe(III) minerals such as poorly crystalline Fe(OH)3 (Supplementary Fig. 1). Bacterial 16S rRNA gene copy numbers increased by four orders of magnitude during these first 125 days of incubation with CH4 (Fig. 4a). While in the original sediment, 16S rRNA gene copy numbers for bacteria and archaea were similar, after 125 days, bacteria dominated over archaea by up to three orders of magnitude and pmoA genes were abundant (particulate methane monooxygenase, used to target aerobic methanotrophs23 as well as nitrite-dependent CH4 oxidation24). This suggests that, since our experiments were anoxic and did not contain other electron acceptors (such as NOx), microorganisms possessing this gene can reduce Fe(III). Furthermore, the mcrA gene, a marker gene commonly used for the quantification of anaerobic CH4-oxidizing archaea25 was present in all CH4-amended microcosms (Fig. 4a).
a Quantitative PCR of bacterial and archaeal 16S rRNA genes, as well as mcrA and pmoA genes before the incubation (Sed T0) and after 125 and 220 days of six parallel incubations with CH4. b Abundance based on DNA-based 16S rRNA gene sequencing. c Active taxa based on RNA-based 16S rRNA sequencing. The presented taxa were analyzed at genus level (and labeled with highest descriptive taxonomic level) and minimum abundance level of 0.5%.
While the original sediments were dominated by archaea belonging to the phylum Thaumarchaeota (22%) and Bathyarchaeia (17%) (Fig. 4b), during the incubation of microcosms supplied with CH4, microorganisms nearly identical (99%) to Methylogaea oryzae jcm 16910 were highly enriched (30–75% relative abundance). In the original sediments and in the biotic and abiotic controls they showed very low abundance (<0.05%). RNA-based analysis (Fig. 4c) showed that M. oryzae was also dominating the active microbial community (30–80%), suggesting its capacity to use CH4 as an electron donor and probably Fe(III) as an electron acceptor. Although M. oryzae was characterized previously as aerobic methane oxidizer26, in our experiment it oxidized methane under Fe(III)-reducing conditions. In its genome (IMG ID: 2675903437), we found over 30 protein-coding genes involved in cytochrome c synthesis and 19 protein-coding genes for Fe-complex membrane receptors and Fe-transport systems (Supplementary Table 2). The genome of M. oryzae jcm 16910 contains a pulE gene and its homolog ferE, which were shown to be involved in Fe(III) reduction in Geobacter metallireducens and Shewanella putrefaciens (Supplementary Table 3). These findings suggest that M. oryzae jcm 16910 and the closely related methanotrophs enriched in our microcosms have the genetic potential to use Fe(III) as an electron acceptor for CH4 oxidation.
Between days 125 and 220, the Fe(III) reduction extent increased significantly (up to 26%) in the CH4-amended microcosms (ANOVA, p < 0.005) (Supplementary Fig. 2). More than 1.5 mg Fe(II)/g sediment was produced (Fig. 3a) and 26 mg/L Fe2+ was released into solution (Fig. 3b). No or only minor (2%) Fe(III) reduction was observed in abiotic CH4-amended and biotic non-CH4-amended microcosms (Supplementary Fig. 2), confirming that in active CH4-amended microcosms, Fe(III) was reduced microbially with CH4 as an electron donor. Dissolved As increased significantly (ANOVA, p < 0.005) to ~3 µg/L (4% of total As). Considering the lower water/sediment ratio in the aquifer (1:8 wt/wt) and a porosity of 0.25 (with a sediment density equal to quartz)7 compared to our microcosms (water/sediment = 3.3:1 wt/wt and porosity of 1), the As concentration in our experiment (3 µg/L) corresponds to 80 µg/L in the field. This suggests that in wells with abundant CH4 (61–509 µg As/L, Fig. 1), substantial amounts of As could be released as a consequence of CH4 oxidation. The calculated CH4 oxidation rate (3.16 × 10−6 mol CH4 cm−3 yr−1) (Supplementary Note 2) is comparable to those measured in other environments (Supplementary Table 4).
Sequences related to M. oryzae that were abundant after 125 days (>30%), decreased in two out of three microcosms after 220 days (to only 0.15 and 1.5% of the total microbial community). Only in one microcosm, M. oryzae-related sequences represented still ~30%. However, in all three CH4-amended microbially active microcosms, another taxon, classified as an archaeon affiliating to Ca. Methanoperedens, became dominant. While its relative abundance before the incubation was only 0.16% and after 125 days it was detected only in one CH4-amended microcosm (0.3%), after 220 days its relative abundance increased to 41–64% in all three CH4-amended microcosms. An increase in archaea was also confirmed by qPCR (Fig. 4a): while bacterial 16S rRNA gene copy numbers, which dominated at day 125, decreased, the archaeal gene copy numbers increased at day 220 by two orders of magnitude compared to bacteria and four orders of magnitude compared to the initial archaeal population. Furthermore, the mcrA gene abundance also increased in all CH4-amended biotic microcosms correlating with the archaeal 16S rRNA gene copy number and implying that the majority of archaea in our microcosms are capable of CH4 oxidation. In contrast, pmoA gene copy numbers decreased, mirroring the disappearance of M. oryzae carrying the pmoA gene. RNA-based sequencing showed that Ca. Methanoperedens was also the most abundant taxon among the active microbial community at day 220 (Fig. 4c), suggesting that it overgrew the initially dominating bacteria related to M. oryzae and that Ca. Methanoperedens was responsible for the increased Fe(III) reduction between days 125 and 220.
The rather low abundance (1–4%) of the known Fe(III)-respiring microorganisms Geobacter sp. and Geothrix sp. (Fig. 4a) suggests that they were only marginally involved in Fe(III) reduction. The experiments were carried out under anoxic conditions, had no (NO3−) present, and accumulation of dissolved Mn2+ and S-species was not observed to a significant extent (Supplementary Fig. 4), which ruled out O2, NO3−, Mn(IV), and oxidized S-compounds as electron acceptors (the pH remained stable at 7.5 ± 0.3 throughout the incubations). This observation sheds new light on the commonly accepted microbially mediated reductive dissolution of Fe(III) minerals and underline importance of methanotrophic archaea in this process.
A co-occurrence of high As, Fe, and CH4 concentrations was reported for many regions of Southeast Asia27. Analysis of >900 groundwater samples across Bengal-, Mekong-, and Red River deltas revealed the highest As concentration in methanogenic zones28 as well as at our field site (Fig. 1). The relationship between high CH4, Fe, and As was usually explained by degradation of organic material via fermentation and methanogenesis creating reducing conditions that lead to reductive dissolution of As rich-Fe(III)-bearing sediments. Our results demonstrated that CH4 is omnipresent and can serve as an electron donor for methanotrophic microorganisms promoting Fe(III)-mineral reduction and dissolution and in consequence contribute to mobilization of As bound to these minerals. In summary, our data suggest that CH4-driven As mobilization can happen in many environments similar to the studied aquifers in Vietnam, where As-bearing Fe(III) minerals are available and CH4 is present. Our study therefore provides compelling new insights into the puzzling mechanisms of As mobilization and groundwater contamination.
Methods
Study area and sample collection
The sampling site is situated close to Van Phuc village, about 15 km SE from Hanoi, on a meander of the Red River (20°55'18.7″N, 105°53'37.9″E) (Fig. 1a). The lithology, geology, mineralogy, characterization, and distribution of organic carbon were described previously17,18,19,29,30,20. Briefly, the North-Western part is characterized by Pleistocene aquifer sands and the groundwater is slightly reduced with As concentrations below the WHO guideline (10 µg/L), whereas the South-Eastern part is of strongly reduced younger gray Holocene sands and the groundwater exceeds the 10 µg/L limit by factors of 10–50 (ref. 19). The transition between the contaminated and uncontaminated zones is characterized by changing redox conditions (redox transition zone) where elevated concentrations of CH4 of up to 45 mg/L were reported20.
In October 2017, a first sampling campaign took place during which we collected a sediment core (ø10 cm; each individual piece ca. 1 m long) from 3.3 to 46.3 m below ground surface at the redox transition zone using rotary drilling (20°55'18.8″ 105°53'38.3″). Samples from several depths along the core were collected for DNA extraction. Groundwater was collected from five wells located in the vicinity of the redox transition zone. Up to 5 L of water pumped directly from the wells was filtered sequentially through 0.8, 0.45, and 0.22 µm pore size filters using filtration holders (Sartorius™ Polycarbonate Filtration Holders). Sediment and filters were kept on dry ice until transported to the laboratory where they were immediately frozen.
In November 2018, a second sampling campaign took place during which another rotary drilling was performed at the same location (3 m away from the 2017 drilling location). For the microcosm setups we chose orange sediments from 40 m depth. Our preliminary data showed that these sediments had high As and Fe contents, and they were the most homogeneous regarding lithology and color (which allowed for sufficient quantities of the representative material for all parallel microcosms). Moreover, these sediments are expected to be responsible for the As release observed at that field site. The sediment was stored anoxically (under N2 atmosphere) at 4 °C in the dark and used immediately after shipping to Germany (ca. 1 week). The total Fe, As, and Mn contents of the sediment were determined by XRF (Bruker, AXS S4 Explorer). The total S and C contents were quantified by a Carbon-Sulphur-Analyzer (CSA 5003, Leybold Heraeus, Germany) and inorganic carbon (TIC) was determined by a Carbon-Water-Analyzer (CWA 5003, Leybold Heraeus, Germany). The organic carbon content (TOC) was calculated by subtracting inorganic carbon from total C.
The TDGP was measured directly in the field using miniRUEDI, a portable mass spectrometer (MS) able to quantify dissolved gas concentrations in water via a continuous flow of the water sample through a membrane contractor. The membrane allows for separation of the gas components in the (ground)water into a surrounding pre-evacuated headspace, where the TDGP is measured prior to separation and measurement of the individual gases. Complete details about the functionality of this instrument can be found in ref. 31. In this campaign, the noble gases He, Ar, Kr, in addition to reactive gases N2, CH4, O2, and CO2 were measured using the miniRUEDI. Calibration for CH4 and CO2 was achieved by using a pre-mixed gas bag containing known quantities of CH4 (1%) and CO2 (1%) in addition to N2 (97%) and H2 (1%).
Microcosm setup
Semi-sacrificial microcosms were set up by mixing 30 g of sediment from 40 m depth (orange sandy Fe- and As-bearing sediments that were suggested to be susceptible to As mobilization) with 100 mL sterile synthetic groundwater medium (modified from ref. 32 without As and Fe in the medium) in glass serum bottles (total volume 250 mL). Prior to the preparation of the microcosms, the pH of the medium was adjusted to 7.3 by bubbling with CO2. The pH was monitored along the experiment and it stayed in the range of 7.2–7.8. All microcosms were prepared in an anoxic glovebox (100% N2), closed with rubber stoppers and aluminum caps. Three different microcosm treatments were prepared: (1) abiotic control (CON−/CH4), i.e. microbial respiratory processes inhibited by amendment with 160 mM sodium azide (NaN3) and headspace exchanged with CH4/CO2 (ratio of 9:1 under the pressure of 1.5 bar); (2) biotic control (CON+), i.e. microbially active but without any amendments and headspace exchanged with N2/CO2 mixture (ratio of 9:1); (3) biotic, i.e. microbially active with the headspace exchanged with CH4/CO2 (ratio of 9:1; under the pressure of 1.5 bar). It has to be noted that the amount of added CH4 was estimated based on the highest observed field concentration. In order to obtain the desired concentration of CH4 in the liquid phase, the amount of necessary volume of the CH4 gas in the headspace was calculated based on Henry’s law. At first, vials were flushed with CH4 for 10 min and pressure equalized to 1 bar. Afterwards, one part of the headspace was withdrawn and replaced with CO2 which gave 9:1 CH4/CO2 under 1 bar pressure. Subsequently, half of the initial CH4 and CO2 volume was injected which resulted in mix of CH4/CO2 in the ratio of 9:1 under 1.5 bar pressure (measured with a portable monometer). With this condition concentration of dissolved CH4 should represent ≈45 mg/L. The microcosms were kept at 26 °C in the dark until analysis (without shaking). At two time points (days 125 and 220), three bottles of each treatment were sacrificed for geochemical analysis and molecular studies.
Geochemical analysis
At each time point (days 0, 8, 35, 50, 65, 80, 95, 110, 125, 165, 180, 195, 210, and 221), 2 mL of slurry and sediment were withdrawn using the syringe and needle (ø 1.20 × 40 mm) under anoxic conditions. Samples were centrifuged at 14,000 r.p.m. for 5 min. One hundred microliters of the supernatant were stabilized in 1 M HCl (to avoid oxidation of Fe(II)) and diluted with HCl if necessary for Fe(II) quantification using the Ferrozine assay. Depending on the Fe concentration the samples were diluted either in 400 or 900 µL of 1 M HCl, resulting in a final HCl concentration of 0.2 or 0.1 M). One milliliter of the supernatant was filtered (0.22 µm, PTFE membrane) and stabilized in 1% HNO3 for As and other elements analysis by ICP-MS (8900, Agilent Technologies, USA). Sediment (0.14 ± 0.03 g wet weight) obtained after centrifugation was digested for 1 h with 1 mL of 6 M HCl. One milliliter of the digests was centrifuged (5 min, 14,000 r.p.m.) and 100 µL of the supernatant was diluted in 1 M HCl. Fe(II) was quantified in triplicate using the Ferrozine Assay33. Statistical differences in As and Fe concentration in the different microcosm setups were analyzed with single factor ANOVA while those at selected time points between pairs of treatments were determined using the Student’s t-test.
Microbial community analysis and quantitative PCR
The DNA from sediment and groundwater samples collected during 2017 sampling campaign as well as the microcosms sediments samples that were collected at the beginning of the experiment, after 10 days (when maximum Fe(III) reduction and As release were observed) and at the end of the experiment (100 days) was extracted following a phenol–chloroform protocol from Lueders et al.34. The RNA was successfully obtained and transcribed for three samples supplied with CH4 (CH4/2, CH4/3, CH4/5). In these samples DNA was digested using the TURBO DNA-free™ Kit, screening PCR and subsequent gel electrophoresis was performed in order to confirm complete removal of DNA. Afterwards, reverse transcription was performed using SuperScript™ III Reverse Transcriptase. Bacterial and archaeal 16S rRNA genes were amplified from DNA and cDNA using universal primers 515f: GTGYCAGCMGCCGCGGTAA35 and 806r: GGACTACNVGGGTWTCTAAT36 fused to Illumina adapters. Subsequent library preparation steps (Nextera, Illumina) and 250 bp paired-end sequencing with MiSeq (Illumina, San Diego, CA, USA) using v2 chemistry were performed by Microsynth AG (Switzerland) and between 45,000 and 242,000 read pairs were obtained for each sample.
16S rRNA (gene) sequence analysis
Sequencing data were analyzed with nf-core/ampliseq v1.1.0 that wraps all analysis steps and software and is publicly available at https://github.com/nf-core/ampliseq37. Briefly, primers were trimmed and untrimmed sequences were discarded (<4%) with Cutadapt v1.16 (ref. 38). Adapter and primer-free sequences were imported into QIIME2 v2018.06 (ref. 39), quality checked with demux (https://github.com/qiime2/q2-demux), and processed with DADA2 v 1.6.0 (ref. 40) to remove PhiX contamination, trim reads (before median quality drops below 35, that is forward 194, reverse 174), correct errors, merge read pairs and remove PCR chimeras, and, ultimately, 6546 amplicon sequencing variants (ASVs) were obtained for groundwater, 2242 for core sediments from 2017, and 4544 from microcosms sediments. Alpha-rarefaction curves were produced with the QIIME2 diversity alpha-rarefaction plugin which indicated that the richness of the samples has been fully observed. A Naive Bayes classifier was fitted with 16S rRNA gene sequences extracted from SILVA v132 (ref. 41) QIIME compatible database 99% identity clustered sequences using the PCR primer sequences. ASVs were classified by taxon using the fitted classifier (https://github.com/qiime2/q2-feature-classifier). ASVs classified as chloroplast or mitochondria were removed. The number of removed ASVs was 70, 9, and 33 for groundwater, core sediment, and microcosm sediments, respectively, totaling to <1% relative abundance per sample and the remaining ASVs had their abundances extracted by feature-table41.
Raw sequencing data have been deposited at DDBJ/ENA/GenBank under BioProject accession number PRJNA593718.
Quantitative PCR
The qPCR specific for the 16S rRNA (genes) of bacteria and archaea as well as for methyl-coenzyme M reductase subunit alpha (mcrA) and particulate methane monooxygenase (pmoA) genes were performed. The qPCR primer sequences, gene-specific plasmid standards, and details of the thermal programs are given in Supplementary Table 1. Total numbers of bacteria and archaea genes were estimated by quantitative PCR (qPCR) (Bio-Rad Laboratories GmbH, Munich, Germany) based on the amplification of the 16S rRNA genes. Quantitative PCRs on DNA extracts obtained as described above were performed in triplicates using SybrGreen® Supermix (Bio-Rad Laboratories GmbH, Munich, Germany) on the C1000 Touch thermal cycler (CFX96TM real time system). Plasmid vectors (pCR2.1®, Invitrogen, Darmstadt, Germany) containing a cloned 16S rRNA gene fragment from Thiomonas sp. and Halobacterium salinarum were used as standards for the quantification of total bacteria and archaea, respectively. Each qPCR assay was repeated three times with triplicate measurements of each sample per run. Data analysis was done using the Bio-Rad CFX Maestro 1.1, software, version 4.1 (Bio-Rad, 2017). Due to low concentrations of extractable DNA in CON+ and CON− sediments after 125 and 220 days, quantification of bacterial and archaeal 16S rRNA genes as well as mcrA and pmoA genes was not successful for these samples.
Data availability
The 16s rRNA amplicon sequencing data is already deposited and publicly available at the NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA593718). The remaining data are submitted to the publicly available repository (www.pangaea.de).
Change history
29 November 2020
A Correction to this paper has been published: https://doi.org/10.1038/s43247-020-00068-5
16 December 2020
A Correction to this paper has been published: https://doi.org/10.1038/s43247-020-00068-5
References
Karagas, M. R., Gossai, A., Pierce, B. & Ahsan, H. Drinking water arsenic contamination, skin lesions, and malignancies: a systematic review of the global evidence. Curr. Environ. Health Rep. 2, 52–68 (2015).
Berg, M., Tran, H. C., Nguyen, T. C., Pham, H. V., Schertenleib, R. & Giger, W. Arsenic contamination of groundwater and drinking water in Vietnam: a human health threat. Environ. Sci. Technol. 35, 2621–2626 (2001).
Charlet, L. & Polya, D. A. Arsenic in shallow, reducing groundwaters in Southern Asia: an environmental health disaster. Elements 2, 91–96 (2006).
Berg, M. et al. Magnitude of arsenic pollution in the Mekong and Red River Deltas—Cambodia and Vietnam. Sci. Total Environ. 372, 413–425 (2007).
Smith, A. H., Lingas, E. O. & Rahman, M. Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ. 78, 1093–1103 (2000).
Lenoble, V., Bouras, O., Deluchat, V., Serpaud, B. & Bollinger, J.-C. Arsenic adsorption onto pillared clays and iron oxides. J. Colloid Interface Sci. 255, 52–58 (2002).
Islam, F. S. et al. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430, 68–71 (2004).
McArthur, J. M. et al. Natural organic matter in sedimentary basins and its relation to arsenic in anoxic ground water: the example of West Bengal and its worldwide implications. Appl. Geochem. 19, 1255–1293 (2004).
Harvey, C. F. et al. Arsenic mobility and groundwater extraction in Bangladesh. Science 298, 1602–1606 (2002).
Islam, F. S. et al. Interactions between the Fe(III)-reducing bacterium Geobacter sulfurreducens and arsenate, and capture of the metalloid by biogenic Fe(II). Appl. Environ. Microbiol. 71, 8642–8648 (2005).
Rowland, H. A. L. et al. The control of organic matter on microbially mediated iron reduction and arsenic release in shallow alluvial aquifers, Cambodia. Geobiology 5, 281–292 (2007).
Mailloux, B. J. et al. Advection of surface-derived organic carbon fuels microbial reduction in Bangladesh groundwater. Proc. Natl Acad. Sci. USA 110, 5331–5335 (2013).
Glodowska, M. et al. Role of in situ natural organic matter in mobilizing as during microbial reduction of feiii-mineral-bearing aquifer sediments from Hanoi (Vietnam). Environ. Sci. Technol. 54, 4149–4159 (2020).
Lapworth, D. J., Gooddy, D. C., Butcher, A. S. & Morris, B. L. Tracing groundwater flow and sources of organic carbon in sandstone aquifers using fluorescence properties of dissolved organic matter (DOM). Appl. Geochem. 23, 3384–3390 (2008).
Anawar, H. M. et al. Mobilization of arsenic in groundwater of Bangladesh: evidence from an incubation study. Environ. Geochem. Health 28, 553–565 (2006).
Al Lawati, W. M. et al. Characterisation of organic matter and microbial communities in contrasting arsenic-rich Holocene and arsenic-poor Pleistocene aquifers, Red River Delta, Vietnam. Appl. Geochem. 27, 315–325 (2012).
van Geen, A. et al. Retardation of arsenic transport through a Pleistocene aquifer. Nature 501, 204–207 (2013).
Weinman, B. The evolution of aquifers and arsenic in Asia: a study of the fluvio-deltaic processes leading to aquifer formation and arsenic cycling and heterogeneity in Bangladesh, Vietnam, and Nepal, Vanderbilt University. https://core.ac.uk/download/pdf/46927487.pdf (2010).
Eiche, E. et al. Geochemical processes underlying a sharp contrast in groundwater arsenic concentrations in a village on the Red River delta, Vietnam. Appl. Geochem. 23, 3143–3154 (2008).
Stopelli, E. et al. Spatial and temporal evolution of groundwater arsenic contamination in the Red River delta, Vietnam: interplay of mobilisation and retardation processes. Sci. Total Environ. 717, 137143 (2020).
Ettwig, K. F. et al. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl Acad. Sci. USA 113, 12792 (2016).
Cai, C. et al. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12, 1929–1939 (2018).
Luesken, F. A. et al. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77, 3877–3880 (2011).
Welte, C. U. et al. Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ. Microbiol. Rep. 8, 941–955 (2016).
Friedrich, M. W. in Methods in Enzymology Vol. 397, 428–442 (Academic Press, 2005).
Geymonat, E., Ferrando, L. & Tarlera, S. E. Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int. J. Syst. Evol. Microbiol. 61, 2568–2572 (2011).
Dowling, C. B., Poreda, R. J., Basu, A. R., Peters, S. L. & Aggarwal, P. K. Geochemical study of arsenic release mechanisms in the Bengal Basin groundwater. Water Resour. Res. 38, 12-1–12-18 (2002).
Buschmann, J. & Berg, M. Impact of sulfate reduction on the scale of arsenic contamination in groundwater of the Mekong, Bengal and Red River deltas. Appl. Geochem. 24, 1278–1286 (2009).
Eiche, E. et al. Origin and availability of organic matter leading to arsenic mobilisation in aquifers of the Red River Delta, Vietnam. Appl. Geochem. 77, 184–193 (2017).
Berg, M. et al. Hydrological and sedimentary controls leading to arsenic contamination of groundwater in the Hanoi area, Vietnam: the impact of iron-arsenic ratios, peat, river bank deposits, and excessive groundwater abstraction. Chem. Geol. 249, 91–112 (2008).
Brennwald, M. S. et al. A portable and autonomous mass spectrometric system for on-site environmental gas analysis. Environ. Sci. Technol. 50, 13455–13463 (2016).
Rathi, B. et al. Processes governing arsenic retardation on Pleistocene sediments: adsorption experiments and model-based analysis: as sorption on Pleistocene sediments. Water Resour. Res. 53, 4344–4360 (2017).
Schaedler, F. A. et al. Revised iron extraction protocol for environmental samples rich in nitrite and carbonate. Geomicrobiol. J. 35, 23–30 (2018).
Lueders, T. et al. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78 (2004).
Parada, A. E. et al. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016).
Apprill, A. et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137 (2015).
Straub, D. et al. Interpretations of microbial community studies are biased by the selected 16S rRNA gene amplicon sequencing pipeline. Preprint at https://www.biorxiv.org/content/10.1101/2019.12.17.880468v1https://www.frontiersin.org/articles/10.3389/fmicb.2020.550420/abstract (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Bolyen, E. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583 (2016).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Acknowledgements
The authors thank all AdvectAs project members for the collaboration and support. Special thanks to Pham Hung Viet, Pham Thi Kim Trang, Vi Mai Lan, Mai Tran and Viet Nga from Hanoi University of Science for the assistance during the sampling campaign. We also thank G. Morin and P. Le Pape from Institut de Minéralogie, de Physique des Matériaux et de Cosmochimie (IMPMC), Paris, for having collected and analyzed XANES data at the As K-edge reported in Supplementary Figure 3. We also thank C. Stengel and N. Pfenninger from Eawag for the technical assistance during ICP-MS analyses. This study was supported by the Deutsche Forschungsgemeinschaft (DFG) (KA 1736/41-1). D.S. is funded by the Institutional Strategy of the University of Tübingen (DFG, ZUK 63) and by the Collaborative Research Center CAMPOS (Grant Agreement SFB 1253/1 2017). S.K. is funded by an Emmy-Noether fellowship (DFG, grant #326028733). The authors acknowledge support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant no INST 37/935-1 FUGG. We acknowledge support by Open Access Publishing Fund of University of Tübingen.
Author information
Authors and Affiliations
Consortia
Contributions
The original hypothesis was formulated by M.G and A.K supported by the field data obtained from A.L and R.K. M.G designed the project, collected the samples interpreted results, and wrote the manuscript with supervision of A.K. E.S collected hydrochemistry data from the field and microcosm experiment and M.S collected data for the solid-phase geochemistry and mineralogy. D.S conducted 16S rRNA sequencing data processing and bioinformatics. S.K and M.J. help to interpret date and provided support with molecular microbiology. M.B. organized sampling campaigned and contributed to field data interpretation. B.R. helped with field data evaluation and geochemical calculations. All AdvectAs members contributed to 2017 and 2018 sampling campaign and all authors contributed to the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary handling editors: Teresa Ortner.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Glodowska, M., Stopelli, E., Schneider, M. et al. Arsenic mobilization by anaerobic iron-dependent methane oxidation. Commun Earth Environ 1, 42 (2020). https://doi.org/10.1038/s43247-020-00037-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-020-00037-y
This article is cited by
-
Prevention of well clogging during aquifer storage of turbid tile drainage water rich in dissolved organic carbon and nutrients
Hydrogeology Journal (2023)
-
An evolving view on biogeochemical cycling of iron
Nature Reviews Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.