Abstract
There is an emerging consensus that microbial necromass carbon is the primary constituent of stable soil carbon, yet the controls on the stabilization process are unknown. Prior to stabilization, microbial necromass may be recycled by the microbial community. We propose that the efficiency of this recycling is a critical determinant of soil carbon stabilization rates. Here we explore the controls on necromass recycling efficiency in 27 UK grassland soils using stable isotope tracing and indicator species analysis. We found that recycling efficiency was unaffected by land management. Instead, recycling efficiency increased with microbial growth rate on necromass, and was highest in soils with low historical precipitation. We identified bacterial and fungal indicators of necromass recycling efficiency, which could be used to clarify soil carbon stabilization mechanisms. We conclude that environmental and microbial controls have a strong influence on necromass recycling, and suggest that this, in turn, influences soil carbon stabilization.
Similar content being viewed by others
Introduction
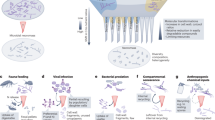
Protecting and increasing soil carbon (C) storage is a global challenge, with ramifications for food security and climate change mitigation1. Historic focus on plant input quantity and quality as key controls on soil C storage has only recently shifted in recognition that plant C assimilation into microbial biomass is an important precursor to soil C stabilization2,3. This relatively unexplored paradigm change in soil C stabilization theory is missing fundamental knowledge. We know that soil microbes assimilate plant C inputs for growth, and release C as extracellular products and CO2 (Fig. 1A): the balance of this anabolism and catabolism is microbial carbon use efficiency (CUE). However, when soil microbes die, microbial residues (hereafter “necromass”) can be recycled as new substrate for microbial growth (Fig. 1B) or stabilized through adsorption to soil mineral surfaces and protection within microaggregates4,5. As necromass forms 50–80% of stable soil organic C6,7,8, it follows that microbial processing of plant-derived C, the absolute amount of necromass production, and recycling of microbial necromass are important controls on soil C storage2,9,10,11,12. We propose that the efficiency of necromass recycling is a critical parameter determining soil C storage, that has not previously been assessed. Quantifying the efficiency of necromass recycling compared with labile plant inputs will provide a more complete picture of a soil’s capacity to stabilize C.
Land use intensity (LUI) is a strong global change driver across terrestrial ecosystems. In temperate agricultural grasslands, higher soil C losses have been detected from soils under intensive management1. LUI is also positively correlated with microbial processes that promote soil C turnover13,14, suggesting that high LUI may promote low microbial CUE. Alternatively, high microbial CUE may offset C losses due to intensive land management (high LUI). Soil microbial communities and activities are strongly shaped by climate and LUI, through impacts on the edaphic environment and the quality and quantity of plant inputs15,16,17. We hypothesized that soil disturbance in high LUI sites (i.e., with tillage, grazing, and fertilization) would select for rapid growth microbes (i.e., “r-selected”) adapted to use labile substrates such as root exudates, and that this would result in higher CUE9,18. In contrast, low LUI sites were predicted to contain more slow growth microbes (i.e., “K-selected”), with the capacity to recycle both labile and complex (e.g., necromass) substrates. We also expected that environmental factors (i.e., soil properties and climate), and land use intensity would interact to influence microbial community composition and resultant necromass recycling.
The aims of the study were to: 1. quantify microbial necromass recycling efficiency in response to land use management intensity, 2. characterize environmental and microbial community controls on necromass recycling efficiency, and 3. identify those microbes associated with high necromass recycling efficiency. To address these aims, we selected nine UK agricultural grassland farms (Fig. 2a) along precipitation, temperature, soil pH, and soil C gradients (Supp. Tables 1 and 2) and identified three LUI levels at each farm creating 27 sites in total. LUI levels were categorized (i.e., high, mid, and low intensity management) and were not consistent across farms (Fig. 2b). Each site was assigned a LUI score calculated from four grassland management activities: mowing (mows y−1), tillage (deep ploughs y−1), nitrogen addition (kg N ha−1 y−1), and grazing (livestock units ha−1 y−1). These factors were averaged over the previous 10 years at each site, standardized across all sites, then added with equal weighting19 (Fig. 2b and Supp. Table 3). We assessed microbial CUE in temperature and moisture-controlled laboratory soil incubations using two substrates: 13C-glucose or microbial necromass. Glucose is a proxy for the sugars that dominate root exudates (~60%) and represents a microbial substrate that can be rapidly assimilated without enzyme catalysis, unlike necromass. The necromass was made from harvesting, then killing, Escherichia coli biomass grown on 13C-labeled glucose. We used stable isotope probing of microbial phospholipids and statistical analyses of 16S- and ITS-sequenced community data to identify microbial guilds, that dominate glucose and necromass uptake and recycling.
Here, we show that soil microbial necromass recycling efficiency is not sensitive to historical land use intensity, but does negatively correlate with historical precipitation (R2adj = 0.53). Microbial carbon use efficiency on both glucose and necromass increases with microbial growth rate on that substrate. We identify 15 bacterial and fungal indicator taxa that have exclusive fidelity and predictive power for necromass recycling efficiency, that can be used in more focused examinations of necromass recycling. The process from necromass carbon production to stable soil carbon has several stages that require further investigation. This study illustrates that the microbial necromass recycling can interrupt that process and is sensitive to precipitation and microbial physiology.
Results and discussion
Historical land use intensity does not alter carbon use efficiency (CUE)
Our CUE-assays on grassland soils across a broad range of historical LUI (Fig. 2b) did not support the hypothesis that LUI was associated with microbial CUE. Using farm as a covariate, neither glucose nor necromass-CUE was responsive to categorical levels of LUI, standardized LUI index score, or individual LUI components (tillage, N fertilizer, mowing or grazing) (P > 0.05; Fig. 3a, c and Supp. Table 5). Although LUI index score independently explained a small amount of glucose-CUE variation (3%, Fig. 4a), this was presumably encompassed within shared variance between LUI and environmental and microbial controls. The location in the UK (farm) did influence the two components of necromass-CUE—substrate-induced CO2 release and C assimilation into biomass (Supp. Tables 4 and 5), suggesting that microbial growth and activity on necromass were responsive to LUI within farms, but the two components typically responded in the same direction, resulting in no net effect on CUE20.
Microbial CUE-glucose (a, b, red) and CUE-necromass (c, d, blue) and their relationship with land use intensity index score (a, c) and soil total carbon (%) in the top 15 cm (b, d). Microbial CUE of both substrates does not vary with index score, or with total soil C for necromass-CUE (P > 0.05, n = 135). There is a positive relationship between CUE-glucose and total soil C (P < 0.0001).
Partitioning of the unique and shared variance between the main environmental and microbial controls on CUE-glucose (a) and CUE-necromass (b). Only significant partitions are shown (P < 0.05). Log-relationship between CUE and C uptake rate (proxy for growth rate) for glucose (c) and necromass (d). Curves are the logarithmic regression and the shaded area is the 95% confidence interval for the CUE~growth rate relationship (Glucose: R2adj = 0.16, P < 0.001; Necromass: R2adj = 0.53, P < 0.001), colored by pH & MAP to show relationship between dominant environmental controls on CUE and growth rate (Glucose~pH: R2adj = 0.14, P < 0.001: Necromass~MAP: R2adj = 0.45, P < 0.001) (n = 135).
Intensive land management disrupts soil carbon cycling21,22 and this was confirmed in our soils: C concentrations decreased with increasing LUI across all sites, both for LUI index score (P = 0.004) and categorical LUI (P = 0.008, Supp. Table 2). CUE is assumed to be a predictor of stable soil C2,4,5,6, yet here CUE was not altered by LUI. We attribute this to the composite nature of LUI and CUE metrics, where individual components may interact to result in no overall change in the final value. Furthermore, necromass-CUE was not correlated with soil %C (Fig. 3d) unlike glucose-CUE (Fig. 3b), consistent with our understanding that labile-C is more likely to form soil C9. We interpret this with caution, however, because soil %C does not always predict stable C pools, especially in surface soils23, and particularly across the broad range of soil quality in this study (Supp. Table 2). Although we reason that necromass recycling must be an important precursor to necromass retention in soil (Fig. 1), and we know that necromass is a large component of bulk soil organic matter8, the efficiency of necromass recycling appears to be primarily an indicator of necromass quality as a substrate, and not predictive of soil %C. This suggests that necromass recycling efficiency influences the pool of necromass available for stabilization, but that other controls (i.e., vegetation feedbacks24, soil redox25, texture26, or necromass N content27) have downstream influences on the stabilization process.
Despite the lack of relationship between CUE and LUI, there were indications that microbial physiology was influenced by individual management interventions, encompassed within the LUI index (Supp. Table 5, see ref. 28). Grazing, fertilizer, and tillage promoted or suppressed C assimilation into biomass and CO2 respiration in tandem, resulting in no net effect on CUE. However, LUI and CUE components may also have interacted with gradients in edaphic factors. For instance, the microbial physiological response to LUI may differ in soils of different pH29. Investigations focused on individual management interventions rather than the composite effect of changing LUI may be more relevant for understanding LUI effects on microbial physiology.
Historical precipitation controls necromass CUE
Our second aim was to investigate controls on necromass-C recycling beyond land use intensity. Climate, soil, and microbial community all contributed to the variance in glucose and necromass-CUE among these UK grassland soils. Specifically, microbial community structure, LUI index scores and three other environmental controls (pH, bulk density, and mean annual temperature (MAT)) all independently explained variance in glucose-CUE (Fig. 4a). Typical of many soil microbial investigations, pH and microbial data shared a large amount of variance for glucose-CUE (28%), confirming the pH-specificity of soil microbial communities15.
Across these grasslands, where mean annual precipitation (MAP) ranges from 568 to 1856 mm y−1, MAP explained the majority of the variance in necromass-CUE. Water availability per se did not control necromass-CUE as we standardized soil moisture during these investigations. Controls on microbial C cycling have been attributed to historical precipitation in other large-scale studies30,31,32, possibly through microbial community selection30. This is consistent with our study, which found independent variance in necromass-CUE explained by both MAP and the bacterial community structure. The best model for understanding controls on necromass-CUE (Fig. 4b) indicated that environment (MAP 55% and bulk density 6%) controlled 61% of the variation, independent of the 3% explained by the 1st bacterial PCOA axis. Our second-best model explained similar independent environmental variance (60%) and more shared microbial-environmental variance (38%), if the 1st fungal PCOA axis is included. This difference between independent and shared microbial variance in the top two models for necromass CUE suggests that environmental and microbial (bacterial and fungal) interactions are also important for necromass-CUE, with a small subset of bacteria independently controlling necromass-CUE across all sites.
In addition to microbial community structure, microbial growth rate on necromass (necromass-C uptake) was strongly negatively correlated with MAP (Fig. 4d). Historically, soil C studies have focused on C losses through microbial respiration instead of microbial growth and turnover. Furthermore, growth was assumed to be low for microbes with high CUE (low progeny per gram substrate33,34,35). Recently reassessed evidence suggests growth to be high for microbes with high CUE9,18,36, consistent with our results. High MAP environments constrain both necromass-CUE (R2adj = 0.53) and microbial growth rate on necromass (R2adj = 0.45). This historical precipitation constraint on C cycling is driven in part by low microbial biomass. Low CUE soils were characterized by a small but potentially active microbial biomass, which led to elevated estimates of qCO2 (biomass-specific respiration) and turnover (biomass-specific substrate-C uptake) at one site (Crianlarich; Supp. Fig. 2a–d). Overall, growth rates were ~10× higher on glucose than necromass, and the glucose: necromass growth rate ratio was also much higher in the high MAP soils (Supp. Fig. 2f), implying a smaller proportion of these microbial communities assimilated necromass as compared to those in low MAP soils. Consequently, soils with low biomass and high growth constraints (i.e., soils with high MAP) have not adapted to use low energy substrates such as necromass with high efficiency.
Microbial guilds of necromass recycling
Our third aim was to identify the soil microbial guilds that are associated with necromass recycling. Specific microbial functional groups were responsible for high uptake of glucose-C and necromass-C, as detected through stable isotope probing of indicative microbial phospholipids (Fig. 5a). There was no differentiation in substrate affinity by farm (Fig. 5b), despite large differences in environmental parameters that control microbial community structure. Together, these results support the theory that substrate is an important determinant of microbial activity at the functional group level17. Substrate affinity for glucose was only partially explained by the dominant environmental control of pH (Fig. 5c), as indicated by the spread in pH along the secondary axis (MDS2) of glucose-C incorporation into microbial phospholipids. For glucose, the specific phospholipid correlational vector arrows are of a similar length, suggesting a common strength in association between substrate and different phospholipids. This broad usage is consistent with our understanding that simple carbon substrate use is shallowly conserved in prokaryotes37. In contrast, necromass-C incorporation into phospholipids was more varied (lipid vector length; Fig. 5d) and showed a pairing between high MAP sites and, in particular, branched chain methyl fatty acids (“GP10Me”) that are typically attributed to actinobacteria38. This is consistent with actinobacteria assimilation of necromass identified in German agricultural Luvisols39.
Microbial substrate use is substrate-specific, not site-specific, as indicated by the clear division by substrate type (a), with no division by site (b). Specific phospholipids responded to glucose (c) and necromass (d), colored by soil pH and mean annual precipitation (MAP), respectively. Vectors represent strength and direction of correlation between individual phospholipids and the axes (n = 45). PLFA-SIP was performed on three LUI levels (low, mid and high) at five farms (n = 3). E.coli-Gram negative peaks (14:0, 16:1ω7, 16:0, 17:1, 18:1ω5, 18:1ω7, and cy-19:0) were removed from the PLFA-SIP analyses of both substrates due to contamination by the necromass addition in the one substrate. Phospholipid precursors indicate functional group: “GP” = Gram+, “GP10Me” = Actinobacteria, “F” = fungi, “U” = unattributed.
To further characterize microbial guilds associated with necromass recycling, we clustered low and high necromass-CUE soils and ran indicator species (IS) analysis on the extant DNA bacterial and fungal operational taxonomic units (OTUs) associated with each cluster. IS are diagnostic of niches or environments and have both high fidelity and predictive value for their cluster, as assessed through multiple permutations40. We found 2306 bacterial and 756 fungal IS that were significant indicators of high necromass-CUE. We filtered this list by assessing the indicator scores of the taxa associated with the IS at multiple taxonomic levels. For instance, our dominant 16S IS was the genus Xiphinematobacter (Verrucomicrobia), a common soil bacterium41. However, this genus also contains species that are common outside of the IS group and so without finer taxonomic resolution is of limited use as an IS. We found nine bacterial and six fungal IS that met our criteria (Table 1). All of the fungal IS and one of the bacterial were at the species level, and validation of the common classes, orders and phyla for the IS suggest that this trait is conserved shallowly. Nonetheless, Actinobacteria had a good fit with high necromass-CUE and MAP (r = 0.46 and −0.57; Table 1), and half of the actinobacterial OTUs were IS for high necromass-CUE. This was surprising as we also found high necromass-C uptake into actinobacterial lipids in high MAP farms (associated with low necromass-CUE) (Fig. 5d), suggesting that preference for necromass substrate by at least some Actinobacteria is not a good indicator of community-level CUE. Therefore, Actinobacteria may be an ideal phyla-level IS for necromass recycling and a less ideal IS for high necromass-CUE. We found the strongest fit with high necromass-CUE and MAP with the Sphingomonodales (r = 0.56 and −0.57, Table 1), a bacterial alphaproteobacterial order containing many low abundance IS. Identification of these 15 IS, the Actinobacteria and the Sphingomonodales in particular, supports our hypothesis that environmental niches can select indicator species for necromass-C recycling in soil.
Our results demonstrate that microbial necromass is a microbial substrate, that necromass-C recycling supports lower microbial growth rates than glucose-C, and that climate and microbial community structure are strongly associated and together control the efficiency of necromass-C assimilation to biomass, versus loss as CO2. The evidence indicates that high microbial growth on necromass and necromass-CUE are found in soils with low historical precipitation. SOM decomposition models that include necromass as a climate-sensitive microbial substrate may assist our prediction of soil C sequestration. Soils with high necromass-CUE are characterized by particular microbial functional groups and their presence and activity will provide diagnostic evidence of sites with efficient necromass recycling. These necromass-recycling diagnostic groups and their climate response provide potential indicators of C sequestration. We conclude that we are only beginning to understand what controls the necromass stabilization process from necromass carbon production to stable soil carbon, and this study describes how microbial recycling can interrupt and is integral to that process. We encourage further research on other stages of this process, such as the rate of biomass turnover to necromass production in different soils, the allocation of necromass to microbial substrate or stable soil organic matter, and the stability of necromass associated with mineral surfaces.
Materials and methods
Site selection and soil sampling
Sites were on a combination of research and commercial farms, on UK agricultural grasslands. We selected nine farms (Supp. Table 1) to span a range in climate and soil types (Supp. Table 2), and interviewed farm managers to characterize land management history (>10 years) (Supp. Table 3) and to identify three land management levels (“sites”) on the same soil type within each farm. In February–March 2016, we visited each farm and established five replicate plots along a transect in each site, with 25 m between each plot. Cores (15 cm × 5 cm diameter) were removed from each plot and stored at 5 °C before and after processing (sieved to 4 mm and adjusted to 70% WHC). We tried to minimize preincubation disturbance but acknowledge that sample handling may reduce sensitivity of the microbial functional responses to historic land use intensity.
Land use intensity index
Site management history (Supp. Table 3) included type and numbers of grazers (converted to DEFRA livestock units ha−1 y−1 42, number of mows and deep ploughs, and amount of fertilizer, slurry or manure applied (converted to kg total N ha−1 y−1). When these numbers varied over time, we calculated the decadal average (2006–2016). We used these data to calculate a land use intensity (LUI) score for each land management level at each site19. This indexing method collapses many complicated site-level variations in an attempt to simplify analysis. It has been used previously to relate LUI to reduced grassland aboveground diversity43 and multifunctionality44.
Isotopically-labeled glucose and necromass
We prepared two substrates: glucose, a proxy for root sugar and a common substrate for CUE assays, and Escherichia coli necromass, a proxy for soil microbial residues. Isotopically labeled glucose solution (20 atm%) was prepared fresh for each carbon use efficiency (CUE) assay by combining 99 atm% glucose with unlabeled glucose; the δ13C value of the glucose solution was calculated by mass for each assay.
Lab-grown E.coli necromass does not represent the biochemical complexity or nutrient content of all soil microbial necromass, yet similar turnover rates on different bacterial and fungal isolates in forest soils45 supported our simplification to a single isolate for this study. Isotopically-labeled E. coli necromass was prepared in advance by growing frozen stock on agar plates, pure cultures in sterile M9 salt solution augmented with filter-sterile 20 atm% 13C-glucose solution for 24 h at 37 °C at 140 rpm, then centrifuging the culture to a single pellet. The pellet cells were lysed with CHCl3, rinsed, sparged, spun down, and autoclaved twice to kill cells. The wet necromass was freeze-dried, and a subsample was suspended in water for each CUE assay to make the necromass substrate, which we assume will contain a mix of cell walls, cell contents and extracellular polymeric substances that were not discarded with the supernatant45. The C content of the necromass powder was measured on a Truspec CN Analyzer (LECO, USA) and the δ13C value of the necromass powder was measured on a ECS-4010 Elemental Analyzer (Costech Analytical Technologies Inc., USA) streamed to a G-2131i cavity ring down spectroscopy (CRDS) isotopic analyzer (Picarro Inc., USA)46. These values and the mass of necromass used in each assay were used to calculate δ13C of the substrate.
Carbon use efficiency assays
We measured growth as the uptake of isotopically-labeled substrate-C into biomass. To examine the effect of simple and complex substrates on carbon use efficiency (CUE), we stabilized soils for 24 h at 15 °C in the dark, then added 1 ml of 13C-necromass or 13C-glucose solutions (20 atm%, 0.05 mg C (g fw soil)−1) each to three microcosms (20 g fw soil each). Two microcosms were for estimating microbial biomass assimilation of substrate into biomass (50 ml tubes), and one microcosm was for measuring CO2 production (250 ml canning jar fitted with gas-tight lid and self-sealing rubber septum). We incubated the microcosms for 24 h in the dark at 15 °C. To estimate cumulative CO2-C production during the incubation, we took four headspace gas samples (5 ml) by syringe and Exetainer® vial (Labco Ltd., UK) and analyzed their CO2 concentration on a Perkin-Elmer Autosystem XL gas chromatograph (GC) fit with a methanizer and flame ionization detector. Our final gas sample coincided with the end of the incubation and a final gas sample, which we used for characterizing the δ13C signature of the headspace gas on a Picarro G2131-i CRDS isotopic analyzer. Simultaneous with the end of the incubation, we extracted the microbial biomass soil using a modified chloroform-direct extraction47, with a water matrix (+1 ml CHCl3 in one of the tubes). Extracts were aliquoted to two streams for estimating extractable carbon and nitrogen, biomass size and substrate incorporation: total organic C analysis on a Shimadzu TOC-L CPN (Shimadzu UK Ltd., UK), and freeze-drying, grinding and 13C analysis on a Costech CN streamed to a Picarro G2131-i CRDS. Reference natural abundance and internal enriched standards were used to calibrate the δ13C analysis for both the gas and solid analysis, and enriched sample δ 13C values (200–8000‰) far exceeded any instrument variability (~0.4–5‰).
Soil and DNA analysis
We measured several soil variables to understand controls on CUE. Soil pH was measured with a HI2211 pH meter (Hanna Instruments Ltd., UK) in a 1:2 (soil:water) slurry. Texture was measured gravimetrically using a hydrometer and cylinder, after removing organic matter (30% H2O2) and carbonates (0.5 M NaOAc). Total C and N were measured on ground samples using a Truspec CN analyzer. Olsen-P was determined colorimetrically48. DNA was extracted from 0.25 g of soil per replicate soil core using PowerSoil-htp 96-well DNA Isolation kit (Qiagen) according to manufacturer’s protocols. A dual indexing protocol49 was used for Illumina MiSeq sequencing of the V3–V4 hypervariable regions of the bacterial 16S rRNA gene using primers 341F50 and 806R51; and the ITS2 region for fungi using primer ITS7f and ITS4r52. Each sample’s amplicon concentrations were normalized using SequalPrep Normalization Plate Kit (Thermo Fisher Scientific) prior to sequencing on the Illumina MiSeq using 600 cycle V3 chemistry. Fungal ITS sequences were processed using PIPITS53 with default parameters. Bacterial sequences were processed using PEAR (sco.hits.org/exelixis/web/software/pear) for merging forward and reverse reads, quality filtered using FASTX tools (hannonlab.cshl.edu), chimera checked with VSEARCH_UCHIME_REF and clustered to 97% OTUs with VSEARCH_CLUSTER (github.com/torognes/vsearch). After processing, the number of amplicon sequence reads per sample was 28205 for 16S rRNA gene and 40406 for ITS2 region.
Calculation of carbon use efficiency
The percentage of CO2-C derived from the added 13C substrates was calculated with a mixing model:
where δC is the δ13C value of the respired CO2 from the control (water only), δT is the δ13C respired CO2 from the substrate-added soils and δS is the δ13C value of the labeled substrate. This value was multiplied by the cumulative CO2-C emissions from the 24 h incubation to calculate the substrate-derived C respired in μg C g−1 soil dwt (“13CO2”).
The percentage of substrate incorporation into microbial biomass flush was determined by:
where MBδ13Cfum is the δ13C value of the microbial biomass from the CHCl3-fumigated soil extract, Cfum is the C pool size of the CHCl3-amended soil water extract (μg C g−1 soil), MBδ13Cunfum is the δ13C value of the soil water extract, and Cunfum is the C pool size of the soil water extract (μg C g−1 soil). The control and treatment values for MBδ13C were incorporated into Eq. (1) to calculate the percentage of substrate incorporation into the microbial biomass (%MB13Csub), then multiplied by the microbial biomass pool size (μg MBC g−1 soil) to convert to mass of substrate incorporated into the biomass (“MB13C”).
Microbial carbon use efficiency was calculated as:
Phospholipid fatty acid stable isotope probing
To quantify substrate utilization by microbial functional groups, phospholipid fatty acids (PLFAs) were extracted from freeze-dried, post-incubation gas microcosm soil54 at the three LUI levels at five farms (n = 3). PLFA peaks were detected on an Agilent Technologies (UK) 6890 GC equipped with a CP-Sil 5CB fused-silica capillary column (50 m × 0.32 mm i.d. × 0.25μm) and flame ionization detector (FID) and identified using a reference library and a GC-MS electron impact ionization spectra on an Agilent Technologies MSD (5973) coupled to a HP6890 GC (same conditions as per GC-FID analysis). δ13C analysis was carried out on fatty acid methyl esters (FAMEs) by Gas Chromatography-combustion-Isotope Ratio Mass Spectrometry (GC-C-IRMS) (Isoprime Ltd). The addition of the extra methyl carbon (methanol −51.14‰) added during the methylation of the fatty acids to their corresponding methyl esters was corrected for accordingly. As above, an isotope mixing model was used to calculate the % of the added substrate recovered in the PLFAs, then this was multiplied by the original PLFA-C pool size to generate substrate-derived PLFAs (ng PLFA-C g−1 soil dw).
Statistical analysis
All statistics were conducted in R (version 3.4.4), with alpha = 0.05 indicating significance. To investigate the response of glucose-CUE and necromass-CUE, 13CO2 and MB13C to LUI, we ran an ANCOVA against the LUI levels and scores and individual LUI component scores, with site as a covariate. To understand the independent and shared variance explained by LUI, environmental and microbial variables on the CUE of glucose and necromass, we used Pearson correlations to isolate significant correlative variables, variance inflation factor analysis (vif function, R package car) and stepwise linear regression (lm and step functions, R package stats), to identify multicollinearity (vif > 10 not retained) and reduce variables (standardized). Retained variables were pooled within the LUI, microbial and environmental categories, and variance partitioning (varpart function in R package vegan) and redundancy analysis (rda function in vegan) was applied to attribute independent and shared variance to LUI, environmental and microbial controls and to assess the significance of these fractions. We used the lm function to assess the log-linear fit of glucose and necromass growth rate against CUE, MAP, and pH. Stable isotope probing into phospholipids (i.e., substrate-derived PLFA-C g−1 dw soil) was visualized in package vegan, using vegdist to make a Bray–Curtis dissimilarity matrix, cmdscale and metaMDS to produce prinicipal coordinates analysis (PCOA) and nonmetric multidimensional scaling (NMDS) ordinations, and envfit to vector correlate lipid scores with samples.
To identify indicator species for necromass recycling, we clustered necromass use efficiency into high-value and low-value clusters with cluster analysis (function kmeans, in stats) and ran indicator species analysis (multipatt, in vegan) on the extant DNA bacterial and fungal operational taxonomic units (OTUs) associated with each cluster. We used multiple permutations (n = 999) to identify both high fidelity and predictive value OTUs for each cluster, and retained bacterial OTUs and fungal OTUs that were significant indicators of high necromass CUE. We assessed the indicator values of the dominant (>0.1% of all OTUs) indicator OTUs at multiple taxonomic levels using Pearson’s correlations. We assessed the response of the indicator OTU relative to the remaining community (proportion > 75%).
Data availability
Data are publicly available: https://doi.org/10.5281/zenodo.3957549
References
Paustian, K. et al. Perspective climate-smart soils. Nature 532, 49–57 (2016).
Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K. & Paul, E. The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Change Biol. 19, 988–995 (2013).
Lehmann, J. & Kleber, M. The contentious nature of soil organic matter. Nature 528, 60–68 (2015).
Miltner, A., Bombach, P., Schmidt-Brucken, B. & Kastner, M. SOM genesis: Microbial biomass as a significant source. Biogeochemistry 111, 41–55 (2012).
Solomon, D. et al. Micro- and nano-environments of carbon sequestration: multi-element STXM–NEXAFS spectromicroscopy assessment of microbial carbon and mineral associations. Chem. Geol. 329, 53–73 (2012).
Kallenbach, C. M., Frey, S. D. & Grandy, A. S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 7, 13630 (2016).
Liang, C. & Balser, T. C. Microbial production of recalcitrant organic matter in global soils: implications for productivity and climate policy. Nat. Rev. Microbiol. 9, 75–75 (2011).
Liang, C., Amelung, W., Lehmann, J. & Kästner, M. Quantitative assessment of microbial necromass contribution to soil organic matter. Glob. Change Biol. 25, 3578–3590 (2019).
Bradford, M. A., Keiser, A. D., Davies, C. A., Mersmann, C. A. & Strickland, M. S. Empirical evidence that soil carbon formation from plant inputs is positively related to microbial growth. Biogeochemistry 113, 271–281 (2013).
Sinsabaugh, R. L., Moorhead, D. L., Xu, X. & Litvak, M. E. Plant, microbial and ecosystem carbon use efficiencies interact to stabilize microbial growth as a fraction of gross primary production. New Phytol. 214, 1518–1526 (2017).
Walker, T. W. N. et al. Microbial temperature sensitivity and biomass change explain soil carbon loss with warming. Nat. Clim. Change 8, 885–889 (2018).
Kallenbach, C. M., Wallenstein, M. D., Schipanksi, M. E. & Stuart Grandy, A. Managing agroecosystems for soil microbial carbon use efficiency: ecological unknowns, potential outcomes, and a path forward. Front. Microbiol. 10, 1146 (2019).
Geisseler, D. & Scow, K. M. Long-term effects of mineral fertilizers on soil microorganisms—a review. Soil Biol. Biochem. 75, 54–63 (2014).
Trivedi, P. et al. Microbial regulation of the soil carbon cycle: evidence from gene-enzyme relationships. ISME J. 10, 2593–2604 (2016).
Griffiths, R. I. et al. The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654 (2011).
Fierer, N. et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl Acad. Sci. 109, 21390–21395 (2012).
Whitaker, J. et al. Microbial carbon mineralization in tropical lowland and montane forest soils of Peru. Front. Microbiol. 5, 720 (2014).
Zheng, Q. et al. Growth explains microbial carbon use efficiency across soils differing in land use and geology. Soil Biol. Biochem. 128, 45–55 (2019).
Blüthgen, N. et al. A quantitative index of land-use intensity in grasslands: integrating mowing, grazing and fertilization. Basic Appl. Ecol. 13, 207–220 (2012).
Hagerty, S. B., Allison, S. D. & Schimel, J. P. Evaluating soil microbial carbon use efficiency explicitly as a function of cellular processes: implications for measurements and models. Biogeochemistry 140, 269–283 (2018).
Guo, L. B. & Gifford, R. M. Soil carbon stocks and land use change: a meta analysis. Glob. Change Biol. 8, 345–360 (2002).
Ward, S. E. et al. Legacy effects of grassland management on soil carbon to depth. Glob. Change Biol. 22, 2929–2938 (2016).
Kramer, M. G., Lajtha, K. & Aufdenkampe, A. K. Depth trends of soil organic matter C:N and 15N natural abundance controlled by association with minerals. Biogeochemistry 136, 237–248 (2017).
Naveed, M. et al. Plant exudates may stabilize or weaken soil depending on species, origin and time. Eur. J. Soil Sci. 68, 806–816 (2017).
Keiluweit, M., Wanzek, T., Kleber, M., Nico, P. & Fendorf, S. Anaerobic microsites have an unaccounted role in soil carbon stabilization. Nat. Commun. 8, 1771 (2017).
Kleber, M., Sollins, P. & Sutton, R. A conceptual model of organo-mineral interactions in soils: self-assembly of organic molecular fragments into zonal structures on mineral surfaces. Biogeochemistry 85, 9–24 (2007).
Kopittke, P. M. et al. Nitrogen-rich microbial products provide new organo-mineral associations for the stabilization of soil organic matter. Glob. Change Biol. 12, 3218–3221 (2017).
Sauvadet, M., Lashermes, G., Alavoine, G. & Recous, S. High carbon use efficiency and low priming effect promote soil C stabilization under reduced tillage. Soil Biol. Biochem. 123, 64–73 (2018).
Malik, A. A. et al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 9, 3591 (2018).
Averill, C., Waring, B. G. & Hawkes, C. V. Historical precipitation predictably alters the shape and magnitude of microbial functional response to soil moisture. Glob. Change Biol. 22, 1957–1964 (2016).
Hawkes, C. V., Waring, B. G., Rocca, J. D. & Kivlin, S. N. Historical climate controls soil respiration responses to current soil moisture. Proc. Natl Acad. Sci. 114, 6322–6327 (2017).
Liu, Z. et al. Precipitation thresholds regulate net carbon exchange at the continental scale. Nat. Commun. 9, 3596 (2018).
Roller, B. R. & Schmidt, T. M. The physiology and ecological implications of efficient growth. ISME J. 9, 1481–1487 (2015).
Shade, A. et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 417 (2012).
Manzoni, S., Jackson, R. B., Trofymow, J. A. & Porporato, A. The global stoichiometry of litter nitrogen mineralization. Science 321, 684–686 (2008).
Manzoni, S. et al. Reviews and syntheses: carbon use efficiency from organisms to ecosystems—definitions, theories, and empirical evidence. Biogeosciences 15, 5929–5949 (2018).
Martiny, J. B. H., Jones, S. E., Lennon, J. T. & Martiny, A. C. Microbiomes in light of traits: a phylogenetic perspective. Science 350, aac9323–aac9323 (2015).
Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils 29, 111–129 (1999).
Apostel, C. et al. Food for microorganisms: position-specific 13C labeling and 13C-PLFA analysis reveals preferences for sorbed or necromass C. Geoderma 312, 86–94 (2018).
Cáceres, M. DE & Legendre, P. Associations between species and groups of sites:\nindices and statistical inference. Ecology 90, 3566–3574 (2009).
Bergmann, G. T. et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 43, 1450–1455 (2011).
Agricultural Budgeting & Costing Book, 81st edn. (Agro Business Consultants Ltd, 2015) https://abcbooks.co.uk/product/abc-budgeting-costing-book-2/.
Gossner, M. M. et al. Land-use intensification causes multitrophic homogenization of grassland communities. Nature 540, 266–269 (2016).
Allan, E. et al. Land use intensification alters ecosystem multifunctionality via loss of biodiversity and changes to functional composition. Ecol. Lett. 18, 834–843 (2015).
Throckmorton, H. M., Bird, J. A., Dane, L., Firestone, M. K. & Horwath, W. R. The source of microbial C has little impact on soil organic matter stabilisation in forest ecosystems. Ecol. Lett. 15, 1257–1265 (2012).
Elias, D. M. O. et al. Functional differences in the microbial processing of recent assimilates under two contrasting perennial bioenergy plantations. Soil Biol. Biochem. 114, 248–262 (2017).
Fierer, N., Allen, A. S., Schimel, J. P. & Holden, P. A. Controls on microbial CO2 production: a comparison of surface and subsurface soil horizons. Glob. Chang. Biol. 9, 1322–1332 (2003).
Emmett, B. A. et al. Countryside Survey. Soils Manual. NERC/Centre for Ecology & Hydrology. 180pp. (CS Technical Report No.3/07 CEH Project Number: C03259) (2008) http://www.countrysidesurvey.org.uk/sites/default/files/CS_UK_2007_TR3%20-%20Soils%20Manual.pdf.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Muyzer, G., Muyzer, G., Smalla, K. & Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Int. J. Gen. Mol. Microbiol. 73, 127–141 (1998).
Yu, Y., Lee, C., Kim, J. & Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005).
Ihrmark, K. et al. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677 (2012).
Gweon, H. S. et al. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980 (2015).
Crossman, Z. M., Abraham, F. & Evershed, R. P. Stable isotope pulse-chasing and compound specific stable carbon isotope analysis of phospholipid fatty acids to assess methane oxidizing bacterial populations in landfill cover soils. Environ. Sci. Technol. 38, 1359–1367 (2004).
Acknowledgements
We thank the farm managers for their time and insights, and Helen Grant and Dafydd Elias for assistance with stable isotope analysis. This work was supported by the Natural Environment Research Council (NERC) Soil Security Programme grants NE/M017125/1 and NE/M016757/1 to R.G., J.W., N.M., and N.O., and an in-kind grant from the NERC Life Sciences Mass Spectrometry Steering Committee (CEH L 105 11 2016) to J.W. for stable isotope analysis.
Author information
Authors and Affiliations
Contributions
K.M.B., N.O., N.M.N., R.G., and J.H. conceived and designed this work; K.M.B., K.M., J.P., T.G., and A.S. acquired the data; K.M.B. interpreted the data and wrote the manuscript; all authors contributed to the revision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary handling editors: Heike Langenberg.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buckeridge, K.M., Mason, K.E., McNamara, N.P. et al. Environmental and microbial controls on microbial necromass recycling, an important precursor for soil carbon stabilization. Commun Earth Environ 1, 36 (2020). https://doi.org/10.1038/s43247-020-00031-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-020-00031-4
This article is cited by
-
Different contributing processes in bacterial vs. fungal necromass affect soil carbon fractions during plant residue transformation
Plant and Soil (2024)
-
Life history strategies of soil bacterial communities across global terrestrial biomes
Nature Microbiology (2023)
-
Linking microbial carbon pump capacity and efficacy to soil organic carbon storage and stability under heavy metal pollution
Soil Ecology Letters (2023)
-
Microbial necromass carbon and nitrogen persistence are decoupled in agricultural grassland soils
Communications Earth & Environment (2022)
-
Life and death in the soil microbiome: how ecological processes influence biogeochemistry
Nature Reviews Microbiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.