Abstract
Bovine serum albumin (BSA) protein is widely used to fabricate antifouling coatings for nanobiotechnology applications. Numerous BSA protein options are commercially available and obtained through different purification methods, however, there is no guidance on which ones are preferable for antifouling coatings. Herein, we investigate the real-time fabrication of antifouling coatings composed of BSA proteins obtained through different purification methods, and report significant differences in ultrathin film coating properties and ability to mitigate serum biofouling and to prevent nanoparticle-induced immune reactions. Mechanistic studies unravel the source of these performance variations, which are related to some BSA proteins containing fatty acid stabilizers while other BSA proteins are fatty acid-free depending on the purification method. Fatty acid-free BSA proteins exhibit greater conformational flexibility and less charge repulsion, which allow them to form more rigidly attached and tightly packed coatings on flat surfaces and nanoparticles that result in superior application performance.
Similar content being viewed by others
Introduction
Ultrathin film coatings are widely used to coat flat, nanostructured, and nanoparticle surfaces in order to confer advantageous properties, such as antifouling or stealth immune profile features1,2. Many types of coating materials, ranging from natural proteins to synthetic nanomaterials such as polymers with finely tuned architectures, have been explored3,4,5 and one of the most popular options for antifouling applications is bovine serum albumin (BSA) protein6. BSA has advantageous properties such as natural abundance, simple coating procedure, low cost, well-established purification methods, and broad availability. Indeed, BSA coatings—often in the form of a single protein monolayer—have long been used in numerous biological assays, including enzyme-linked immunosorbent assays (ELISA)7, blots8, immunohistochemistry9, and polymerase chain reaction10, in order to minimize nonspecific binding events and thus enhance assay performance. In recent years, BSA has also found extensive usage as an antifouling coating material in biosensing11 and nanomedicine12 applications. For example, the simplicity of forming BSA-based coatings has inspired the development of advanced antifouling coatings that enable high-performance electrochemical biosensing in complex biological fluids13. In addition, BSA coatings are used to improve nanoparticle stability and biocompatibility14,15, and to passivate surfaces to inhibit nonspecific adsorption of bioanalytes4.
An often-overlooked aspect of BSA is that there are many commercially available versions which differ in how they were isolated from bovine plasma16, even for those from the same company17. Guidelines to predict which BSA version would be most suitable for antifouling applications are lacking despite broad interest in developing a nanodescriptor index to predict protein adsorption at nano-bio interfaces18. To date, most related studies have only evaluated a single BSA version and compared BSA adsorption and surface-induced conformational changes on different surfaces6,19 or investigated the effects of isolated parameters such as ionic strength20 or heating21. However, the comparison of BSA proteins obtained through different purification methods remains unexplored despite being the greatest source of variability. As such, the selection of BSA protein type is largely a matter of trial-and-error and laboratory precedent, and there is oftentimes limited reporting on which particular BSA was used in scientific publications.
Herein, we scrutinize the fabrication of BSA-based antifouling coatings from BSA proteins obtained through different purification methods, and comprehensively investigate BSA conformational and adsorption properties on flat and nanoparticle surfaces in order to identify key mechanistic factors that drive the formation of high-performance ultrathin film coatings with superior antifouling properties.
Results
Evaluation strategy
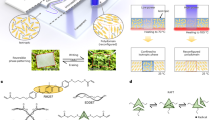
BSA is widely used to form surface passivation coatings on bulk (e.g., glass coverslip) and nanostructured (e.g., inorganic nanoparticles) surfaces. In practice, the surface is incubated with an aqueous BSA solution, whereby protein molecules noncovalently adsorb and undergo a conformational change (“denature”) on the surface to form an ultrathin protein monolayer coating (Fig. 1a). Coating performance is sensitive to various material parameters such as atomic composition22, surface roughness23, and nano-curvature effects24, along with environmental parameters such as solution pH25 and ionic strength20.
a Overview of basic steps in BSA coating protocols. (i) A target surface is identified to coat with BSA for surface passivation. (ii) BSA proteins in aqueous solution are incubated with the target surface. (iii) BSA proteins adsorb and denature on the target surface. (iv) A washing step is performed to remove weakly bound BSA protein molecules. (v) A firmly attached monolayer of adsorbed BSA protein molecules is formed on the surface. b Overall experimental strategy to analyze fabrication of BSA-based ultrathin film coatings in terms of solution-phase protein structure and adsorbed protein layer properties. c Outline of purification routes to isolate BSA protein molecules from bovine plasma. Fatty acid-containing BSA proteins are designated as BSA 1, 2, or 3 depending on the purification method. Fatty acid-free versions of BSA 1, 2, and 3 were also prepared and are designated as BSA 4, 5, and 6, respectively.
Our evaluation strategy focused on characterizing the conformational, adsorption, and antifouling properties of six BSA proteins purified through different methods (Fig. 1b). The six BSA types were purified through three different fractionation routes, without or with a fatty acid removal step (Fig. 1c and Supplementary Table 1). The fractionation step is necessary to separate BSA from other serum components and there are two main options: cold ethanol fractionation and heat-shock fractionation. Briefly, cold ethanol fractionation involves reducing the sample temperature to approximately −5 °C followed by the addition of ethanol and adjusting the solution pH and ionic strength to isolate BSA protein26. Conversely, heat-shock fractionation involves heating the sample (typically to ~60 °C) to isolate BSA protein and is conducted in the presence of a fatty acid stabilizer (caprylic acid)16,27,28,29. These two fractionation processes can be completed alone or sequentially (cold ethanol, then heat-shock).
Notably, after the fractionation step(s) is completed, the purified BSA proteins still contain fatty acids that were either naturally bound or were added as a stabilizer30. We tested fatty acid-containing (“fatted”) BSA proteins prepared by (1) cold ethanol fractionation, (2) heat-shock fractionation, and (3) cold ethanol fractionation followed by heat-shock fractionation and these proteins were designated as BSA 1, 2, 3, respectively. To remove fatty acids, an extra processing step is required, which involves the addition of activated charcoal followed by adjusting solution pH and temperature31. We also tested the fatty acid-free (“defatted”) versions of BSA 1, 2, 3, designated as BSA 4, 5, 6, respectively. Infrared spectroscopic characterization—through the analysis of relevant spectral features—confirmed the removal of fatty acids from defatted BSA proteins, which is in line with gas chromatography results (Supplementary Fig. 1 and Supplementary Note 1).
Conformational stability of fatted and defatted BSA proteins
Like other globular proteins, BSA folds in aqueous solution to minimize its conformational free energy32. The folded structure imparts conformational stability and we first investigated the relative degree of conformational stability of the different BSA proteins by dynamic light scattering (DLS) and circular dichroism (CD) spectroscopy (Fig. 2a). The experiments were initially conducted at 25 °C and then repeated at higher temperatures in 5 °C increments. DLS detected temperature-induced loss of conformational stability when protein molecules irreversibly unfold and begin to aggregate33 and a higher onset temperature indicates greater conformational and/or colloidal stability. CD spectroscopy further evaluated corresponding changes in the secondary structure of protein molecules.
a Schematic illustration of DLS and CD spectroscopy measurement strategies. b Hydrodynamic diameter of BSA protein molecules as a function of temperature, as measured in DLS experiments. Data are reported as mean ± standard deviation (s.d.), where s.d. is defined as the full-width-at-half-maximum (FWHM)/2.355 [n = 5 technical replicates, one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (versus data at 25 °C)]. Dots represent individual data points. c Fractional percentage of α-helicity in BSA protein molecules as a function of temperature, as measured in CD spectroscopy experiments. Mean values are presented on top of each column. Values were computed from molar residue ellipticity data and data are reported as mean ± s.d. (n = 3 technical replicates). Dots represent individual data points. The grey horizontal line in each graph corresponds to 50% helicity.
Across the temperature range of 25 to 55 °C, the DLS data showed that all six BSA proteins have ~8 nm diameter, which agrees well with the expected size of BSA monomers20,21 (Fig. 2b and Supplementary Fig. 2). At 60 °C, a marked increase in the sizes of defatted BSA proteins (4-6) was detected, with average diameters around 20 nm that indicated the onset of protein aggregation. By contrast, the sizes of fatted BSA proteins (1–3) remained stable at 60 °C. At 65 °C, BSA 4–6 increased in size to around 45 nm diameter and the onset of protein aggregation occurred for BSA 1, with an average diameter of around 35 nm. By contrast, the onset temperatures for BSA 2 and 3 were 70 °C and 75 °C, respectively. Kinetic experiments further verified that defatted BSA proteins underwent more rapid and extensive aggregation, which points to fatty acid molecules conferring BSA proteins with greater conformational stability and/or colloidal stability (Supplementary Fig. 3).
In addition, the CD spectroscopy data showed that all six BSA proteins exhibited around 60-63% α-helicity at 25 °C, followed by a progressive decrease in α-helicity with increasing temperature (Fig. 2c and Supplementary Figs. 4, 5). Both reversible (below the onset temperature of aggregation) and irreversible (at and above the onset temperature) conformational changes contribute to protein unfolding so we compared the helicity data at 65 °C. All three defatted BSA proteins had helical percentages below 50% on average, while all three fatted BSA proteins had helical percentages above 51% on average (Supplementary Table 2). More pronounced differences in helicity were observed at 70 °C, and the findings support that, among the fatted BSA proteins, BSA 1 had the lowest α-helicity and hence, lowest conformational stability. The relatively low conformational stability of BSA 1 is consistent with cold ethanol fractionation, whereby only natural fatty acids remain bound (i.e., no additional fatty acid stabilizer). Collectively, the DLS and CD spectroscopy data support that fatted BSA proteins had greater conformational stability than defatted BSA proteins.
Adsorption behavior of fatted and defatted BSA proteins
We then investigated the adsorption behavior of BSA proteins on silica surfaces (Fig. 3a). The experiments were conducted using a BSA protein concentration of 100 µM (~6.6 mg mL−1) in an aqueous buffer (10 mM Tris [pH 7.5] with 150 mM NaCl) which is representative of the typical solution conditions used in BSA coating protocols8,19,34. When a protein with lower conformational stability adsorbs onto a solid surface, it undergoes greater unfolding and spreading on the surface due to entropic gains and other factors such as protein-surface and protein-protein interactions can also affect the outcome35,36. We conducted quartz crystal microbalance-dissipation (QCM-D) measurements to track the adsorption kinetics and corresponding mass and viscoelastic properties of adsorbed BSA protein molecules. The QCM-D technique measures real-time changes in the resonance frequency (ΔF) and energy dissipation (ΔD) signals of an oscillating, silica-coated sensor surface, providing insight into the hydrodynamically-coupled mass and viscoelastic properties of adsorbed BSA protein molecules on the silica surface, respectively.
a Overview of experimental protocol. b–e QCM-D experiments tracking frequency (ΔF) and energy dissipation (ΔD) shifts related to BSA adsorption. b Time-resolved QCM-D ΔF shifts and (c), corresponding |ΔFmax| shifts at saturation. d Time-resolved QCM-D ΔD shifts. e |ΔFmax/ΔDmax| ratios obtained from saturation data in panels b and d. f–g LSPR experiments were conducted to measure wavelength shifts (Δλ) related to BSA adsorption. f Time-resolved LSPR Δλ shifts and (g), corresponding Δλmax shifts at saturation. Data in (c, e, g) are reported as mean ± s.d. (n = 3 biological replicates, one-way ANOVA with Tukey’s multiple comparisons test). P values are reported for defatted BSA proteins (versus fatted BSA proteins from the same fractionation method). Dots represent individual data points.
All six BSA proteins adsorbed onto the silica surface and the QCM-D ΔF shifts indicated monotonic adsorption kinetics (Fig. 3b). The buffer washing step revealed that most adsorbed protein molecules were bound irreversibly. Generally, larger |ΔFmax| shifts around 35 to 45 Hz were observed for defatted BSA proteins (4-6), indicating greater adsorption, compared to 15 to 35 Hz for fatted BSA proteins (1–3) (Fig. 3c). The corresponding QCM-D energy dissipation signals indicated some degree of viscoelastic character within the protein adlayers (Fig. 3d). Further analysis of |ΔFmax/ΔDmax| ratios and time-independent frequency-energy dissipation (F–D) curves revealed that adsorbed, defatted BSA proteins underwent more surface-induced denaturation, as indicated by lower energy dissipation shifts relative to the frequency shift, which is consistent with lower solution-phase conformational stability (Fig. 3e and Supplementary Fig. 6). Conversely, fatted BSA proteins exhibited greater viscoelasticity, indicating less surface-induced denaturation in the adsorbed state.
To corroborate the QCM-D results and extend the results to nanostructured surfaces37, we performed localized surface plasmon resonance (LSPR) experiments to measure BSA protein adsorption onto silica-coated gold nanodisk array surfaces. The LSPR technique detects the wavelength shift (Δλ) that is associated with changes in local refractive index near the sensor surface and the measurement signal is sensitive to the adsorbed protein mass and degree of adsorption-related protein spreading38. For all six BSA proteins, a positive monotonic Δλ shift occurred upon adsorption and most protein molecules remained bound irreversibly after buffer washing (Fig. 3f). Importantly, the Δλmax shifts showed that defatted BSA proteins exhibited greater adsorption uptake than fatted BSA proteins (~1.2 ± 0.2 nm versus ~0.6 ± 0.2 nm), which supports the QCM-D data (Fig. 3g). Thus, defatted BSA proteins exhibited higher adsorption uptake and tighter adlayer packing compared to their fatted counterparts.
Adsorption-related protein conformational changes
To compare the extent of surface-induced protein denaturation for each BSA protein, we further analyzed the LSPR-tracked initial rate of adsorption uptake, denoted as (dΔλ/dt). A larger initial rate is indicative of greater denaturation38,39,40. The LSPR measurements yielded (dΔλ/dt)max values of around 0.6–0.7 nm min−1 for defatted BSA proteins, compared to around 0.2–0.3 nm min−1 for fatted BSA proteins which indicates more extensive surface-induced denaturation for defatted BSA proteins (Fig. 4a and Supplementary Fig. 7).
a Maximum rate of change in the LSPR wavelength shift (dΔλ/dt)max during the initial adsorption stage. Values are computed from data in Fig. 3f. Data are reported as mean ± s.d. (n = 3 biological replicates, one-way ANOVA with Tukey’s multiple comparisons test). b Fractional percentage of α-helicity in BSA protein molecules in solution and in the adsorbed state, as determined in ATR-FTIR spectroscopy experiments. Data are reported as mean ± s.d. (n = 3 biological replicates, two-way ANOVA with Tukey’s multiple comparisons test). P values are reported for defatted BSA proteins (versus fatted BSA proteins from the same fractionation method) in (a), and are separately reported for defatted BSA proteins in solution (versus fatted BSA proteins in solution from the same fractionation method) and for defatted BSA proteins in the adsorbed state (versus fatted BSA proteins in the adsorbed state from the same fractionation method) in (b). Dots represent individual data points.
In addition, changes in BSA protein structure due to adsorption-related conformational changes were measured by attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy. Generally, in the solution phase, the α-helicities of fatted and defatted BSA proteins were around 63% and 60%, respectively, agreeing with literature values22 (Fig. 4b, Supplementary Fig. 8, and Supplementary Table 3). Upon adsorption, fatted BSA proteins underwent surface-induced denaturation and the fractional helicities decreased to around 53% (i.e., a net helical loss of ~10%). By contrast, defatted BSA proteins underwent greater surface-induced denaturation, resulting in fractional helicities around 45%, (i.e., a net helical loss of ~15%). The adsorption-related decrease in α-helicity was related to protein unfolding, as indicated by a corresponding increase in the random coil fraction22,41,42. Specifically, fatted and defatted BSA proteins experienced an increase in random coil fraction by ~9% and ~12%, respectively, supporting that fatty acids partially stabilize BSA proteins against surface-induced denaturation by reducing the extent of the helix-to-random coil secondary structure transition (Supplementary Note 2). Thus, multiple lines of experimental data support that defatted BSA proteins exhibit greater surface-induced denaturation than fatted BSA proteins. This finding is consistent with the lower solution-phase conformational stability of defatted BSA proteins, and supports that decreased solution-phase conformational stability translates into more pronounced adsorption-related denaturation on both flat and nanostructured surfaces.
Antifouling coating performance
We next investigated the application performance of the six BSA proteins in surface-based and nanoparticle-based assays related to antifouling properties. We first measured the surface passivation efficiency of adsorbed BSA coatings to inhibit serum biofouling on silica surfaces (Fig. 5a and Supplementary Fig. 9). A bare silica surface was first coated with BSA protein molecules before incubation in 100% fetal bovine serum (FBS) for 80 min, followed by a washing step. The resulting amount of serum biofouling on the BSA-coated silica surface was evaluated by the QCM-D technique. The passivation efficiency of the BSA coating was determined from their ability to inhibit serum biofouling. We found that defatted BSA proteins outperformed fatted BSA proteins, with defatted BSA 5 and 6 exhibiting ~90% passivation efficiency (Fig. 5b). By contrast, fatted BSA 1 and 3 demonstrated less than 40% passivation efficiency. These findings are consistent with greater adsorption uptake and higher packing density of adsorbed, defatted BSA protein molecules.
a Schematic illustration of serum biofouling assay. Pristine and BSA-coated silica surfaces were incubated in 100% fetal bovine serum (FBS) followed by a buffer washing step, and the amount of serum biofouling was measured. The passivation efficiency of the BSA coatings was calculated relative to the uncoated surface. b Passivation efficiencies of BSA coatings to inhibit serum biofouling (n = 3 biological replicates). c Schematic illustration of nanoparticle-triggered complement activation assay. Pristine and BSA-coated silica nanoparticles were incubated in 100% normal human serum (NHS), followed by ELISA measurements to determine the extent of nanoparticle-triggered complement activation. The degree of protection for BSA-coated nanoparticles was calculated relative to uncoated nanoparticles. d Protection efficiencies of BSA coatings to inhibit nanoparticle-induced complement activation (n = 5 biological replicates). Data in (b) and (d) are reported as mean ± standard error of the mean (s.e.m.) (one-way ANOVA with Tukey’s multiple comparisons test). P values are reported for defatted BSA proteins (versus fatted BSA proteins from the same fractionation method). Dots represent individual data points.
In addition, we tested the surface passivation (“blocking”) performance of fatted and defatted BSA proteins in Western blot experiments involving a hydrophobic nitrocellulose membrane surface (Supplementary Figs. 10–12 and Supplementary Note 3). We first exposed the membrane surface to human serum by electrophoretic transfer, “blocked” the remaining unexposed surface with BSA proteins, and then incubated the membranes with a primary C3 monoclonal antibody followed by a relevant secondary antibody before the intensities of specific and nonspecific bands—produced by enhanced chemiluminescence—were quantified. We found that the membranes “blocked” by defatted BSA proteins produced lower intensity nonspecific bands, supporting that the higher packing density of adsorbed defatted BSA proteins more effectively prevented nonspecific interactions between the primary and/or secondary antibodies and the membrane surface. Together, these data support that defatted BSA proteins exhibit superior surface passivation performance on hydrophilic and hydrophobic surfaces.
We also tested the antifouling coating performance of BSA coatings on silica nanoparticles to inhibit nanoparticle-induced complement activation, which is an innate immune reaction43,44,45 (Fig. 5c). Using ELISA, we measured the degree of protection BSA coatings conferred against nanoparticle-induced complement activation in human serum, as assessed by the amount of SC5b-9 protein biomarker in solution (Fig. 5d and Supplementary Fig. 13). We found that defatted BSA coatings inhibited more than 60% of SC5b-9 generation, whereas the fatted BSA coatings inhibited ~40% or less. This finding supports that defatted BSA coatings are superior to fatted BSA coatings in protecting against nanoparticle-induced complement activation.
Discussion
Our findings demonstrate that defatted BSA proteins exhibit distinct conformational and adsorption properties compared to fatted BSA proteins, and these differences lead to significant variations in antifouling coating performance. Overall, the data highlight the central role of fatty acid molecules in affecting BSA structure and function in the adsorbed monolayer state. To verify the effect of fatty acids, we doped defatted BSA with caprylic acid, thereby converting the defatted protein into a fatted protein. Caprylic acid addition led to markedly enhanced conformational stability, as well as decreased adsorption uptake and surface-induced denaturation (Supplementary Figs. 14–24, Supplementary Tables 4–5, and Supplementary Note 4). Mechanistically, the enhanced conformational stability of fatted BSA proteins can be understood through the insertion of fatty acid molecules (via hydrophobic tails) into hydrophobic pockets on the molecular surface of BSA proteins, conferring a net stabilizing effect on the protein structure46,47 (Fig. 6a).
a Schematic illustration of fatted and defatted BSA protein molecules in solution. b Schematic illustration of fatted and defatted BSA protein molecules in the adsorbed state. Higher conformational stability and charge repulsion of fatted BSA leads to less adsorption-induced denaturation, greater protein-protein repulsion, and thus less tightly packed protein coatings. Conversely, the lower conformational stability and charge repulsion of defatted BSA leads to greater adsorption-induced denaturation, less protein-protein repulsion, and thus more tightly packed protein coatings. As a result, defatted BSA proteins form superior antifouling coatings.
A less obvious but critically important effect of fatty acids on BSA relates to the protein adsorption process itself (Fig. 6b). Generally, a protein undergoes surface-induced denaturation upon adsorption, the extent of which depends on the protein’s conformational stability and strength of protein-surface interactions35. More denaturation causes greater surface spreading and consequently, a larger protein adsorption footprint. Therefore, greater surface spreading—whether due to conformational stability48 or protein-surface interactions (e.g., as mediated by ionic strength effects20,49)—typically results in a smaller total number of adsorbed protein molecules on a target surface.
This scenario holds true for comparing defatted and fatted BSA proteins, however, the difference between the two BSA types is even more pronounced because the fatty acid molecules bound to BSA not only enhance conformational stability but also increase the negative surface charge of BSA protein molecules due to the exposed carboxylic acid headgroups. Thus, while fatted BSA proteins already have a smaller adsorption footprint due to greater conformational stability, the maximum surface density of adsorbed protein molecules in the fatted BSA case is further limited by more repulsive electrostatic interactions between nearest-neighbor protein molecules. These findings support that bound fatty acids enhance not only BSA conformational stability but also charge repulsion between protein molecules in the adsorbed state. As such, defatted BSA proteins are better suited to form passivation coatings since they have lower conformational stability, permitting greater denaturation and spreading in the adsorbed state, and the absence of fatty acids also permits more favorable protein-protein interactions that support the formation of tightly packed, adsorbed protein adlayers.
Taken together, our findings support that it is preferable to use fatty acid-free BSA proteins to form antifouling coatings on flat and nanostructured surfaces. In general, fatty acid-free BSA proteins outperformed fatty acid-stabilized BSA proteins regardless of the fractionation route used in the purification stage. Mechanistically, we show that defatted BSA proteins are superior because they have lower solution-phase conformational stability and reduced charge repulsion in the adsorbed state, which translates into greater surface-induced denaturation and adsorption uptake, resulting in tighter adlayer packing that yields superior passivation coatings.
Looking forward, these findings will be relevant for studying other fatty acid-binding proteins such as human serum albumin across various applications, including protein corona formation, and can also be further explored in the context of nanoparticle properties (i.e., surface chemistry, shape, and size)22,42,50,51. We anticipate that the rational selection of BSA protein options without or with fatty acid stabilizers can enable the fabrication of superior antifouling coatings for a wide range of applications, which can be readily implemented by researchers across different fields of materials science and nanobiotechnology.
Methods
Bovine serum albumin proteins
The six types of bovine serum albumin (BSA) proteins used in this study were selected based on three different fractionation methods without or with a fatty acid removal step, and were procured from Sigma-Aldrich (St. Louis, MO, USA). Lyophilized powders of fatty acid-containing BSA proteins purified by cold ethanol fractionation, heat-shock fractionation, and cold ethanol followed by heat-shock fractionation were selected and have catalog nos. A2153, A3059, and A7638, respectively. These three fatted BSA proteins were labeled BSA 1, 2, 3, respectively. The corresponding fatty acid-free versions were also obtained and have catalog nos. A6003, A7030 and A0281, respectively. These three defatted BSA proteins were labeled BSA 4, 5, 6, respectively. All six BSA proteins were used as provided and the fatty acid-free versions were confirmed to have ≤0.01% fatty acid residues by gas chromatography experiments according to the manufacturer (Supplementary Table 1).
Reagents
Sodium dodecyl sulfate (SDS, catalog no. L4390), sodium chloride (NaCl, catalog no. 746398), sodium hydroxide (catalog no. S5881) and octanoic acid (caprylic acid, catalog no. C2875) were also purchased from Sigma-Aldrich while Tris(hydroxymethyl)aminomethane (Tris, catalog no. 0497) was purchased from Amresco (Solon, OH, USA). Ethanol (95%) was purchased from Aik Moh (Singapore), hydrochloric acid (HCl, catalog no. 100317) was purchased from Merck (Burlington, MA, USA) and fetal bovine serum (FBS, catalog no. SV30160.03, lot no. RC35960) was purchased from HyClone Laboratories (Logan, UT, USA) and stored at −20 °C until experiment. Normal human serum (catalog no. NHS, lot no. 38) was obtained from Complement Technology (Tyler, TX, USA) and stored at −80 °C until experiment. 30% Acrylamide/Bis Solution 29:1 (catalog no. 1610156), ammonium persulfate (catalog no. 1610700), tetramethylethylenediamine (TEMED, catalog no. 1610800), 4× Laemmli sample buffer (catalog no. 1610747), 2-mercaptoethanol (catalog no. 1610710), 10× Tris buffered saline (TBS, catalog no. 1706435), 10× Tris/glycine buffer (catalog no. 1610734), 10× Tris/glycine/SDS buffer (catalog no. 1610732), Tween 20 (catalog no. 1706531), nitrocellulose membranes (catalog no. 1620112), Precision Plus Protein Standards (catalog no. 1610375), Clarity Max Western enhanced chemiluminescent (ECL) substrate (catalog no. 1705062), and horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (catalog no. 1721011, batch no. 64109318) were all purchased from Bio-Rad Laboratories (Hercules, CA, USA). Complement C3b monoclonal antibody (catalog no. MA1-40155, lot no. SH2428445) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Methanol (catalog no. M/4000/17) was obtained from Fisher Scientific (Loughborough, UK) and 100-nm diameter silica nanoparticles (catalog no. SISN100) were obtained from nanoComposix (San Diego, CA, USA).
Sample preparation
An aqueous buffer solution of 10 mM Tris, 150 mM NaCl, and pH 7.5 was prepared with Milli-Q-treated water (resistivity of >18.2 MΩ.cm at 25 °C) and filtered through a 0.2 µm polyethersulfone (PES) membrane filter (Thermo Fisher Scientific, catalog no. 595-4520). BSA solutions were prepared by dissolving lyophilized BSA powder in this Tris buffer and then filtering through a 0.2 µm syringe filter (catalog no. PN-4612; Pall Corporation, Port Washington, NY, USA). The molar concentrations of BSA proteins in aqueous buffer solution were determined by ultraviolet (UV) light absorption measurements at 280 nm and with a molar extinction coefficient value of 43,824 M−1 cm−1. Where applicable, the addition of caprylic acid to BSA samples was conducted by first dissolving caprylic acid and sodium hydroxide into the Tris buffer in order to make a 50 mM caprylic acid solution at pH 7.5. The caprylic acid solution was then added to appropriate amounts of BSA solution in order to yield caprylic acid-doped BSA sample with a molar ratio of 10:1 caprylic acid:BSA. In addition, the Western blot running buffer was prepared by diluting 10× Tris/glycine/SDS buffer with water to 1× concentration, and the transfer buffer was prepared by diluting the 10× Tris/glycine buffer with water and methanol to 1× concentration and 20% (vol/vol) of methanol. The BSA-containing surface passivation (“blocking”) solutions for Western blot experiments were prepared by first diluting 10× TBS with water, followed by the addition of Tween 20 to make a 1× TBS solution with 0.1% (vol/vol) Tween 20 (TBST). BSA was then dissolved in TBST to make a 3% (wt/vol) BSA blocking solution.
Dynamic light scattering
Dynamic light scattering (DLS) was employed to measure the average hydrodynamic diameter and polydispersity of 451 µM (~30 mg mL−1) BSA protein molecules in solution using a particle size analyzer (ZetaPALS, Brookhaven Instruments, Holtsville, NY, USA) that is equipped with a 658.0 nm monochromatic laser. Measurements were taken at a 90° scattering angle and the BIC Particle Sizing software (v5.27; Brookhaven Instruments) was used to analyze the intensity autocorrelation function in order to obtain the intensity-weighted Gaussian size distribution. The temperature in the measurement chamber was controlled with a feedback loop and measurements of a protein sample were first recorded at 25 °C, followed by heating and increasing the temperature from 50 °C to 75 °C in 5 °C increments. After each temperature step increase, the measurement chamber was equilibrated for 5 min before the DLS measurement was performed. In select time-dependent measurements, the temperature in the measurement chamber was first raised from 50 °C to 55 °C before maintaining a constant temperature at 60 °C and measuring protein size every 10 min for 200 min. All reported values were obtained from 5 technical replicates.
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy experiments were conducted using an AVIV Model 420 CD spectrometer with the AVIV CDS software package (v3.36 MX) (AVIV Biomedical, Lakewood, NJ, USA). CD spectroscopy measurements were conducted using 400 µL solutions of 2.5 µM BSA samples in a 1 mm path length cuvette with a PTFE stopper (catalog no. 110-QS; Hellma, Müllheim, Germany). The measurements were recorded with a 1 nm spectral bandwidth, 0.5 nm step size, and an averaging time of 0.1 s. Elevated temperature experiments were conducted by increasing the temperature in the measurement chamber in 5 °C increments from 50 °C to 75 °C and recording the spectra at every temperature point after a 5 min equilibration time. All resulting spectra were processed by subtracting a background spectra of equivalent solution conditions without BSA protein and the data were presented in mean residue molar ellipticity ([θ]) units based on the following equation:
where n is the number of amino acid residues, c is the protein molar concentration, and l is the cuvette path length in cm. All CD spectra were smoothed by using the Savitzky-Golay52 smoothing function with a smoothing window of 20 points and a polynomial order of 2 in the OriginPro 2019b (v9.6.5.169) software package (OriginLab, Northampton, MA, USA). The α-helical percentage of each BSA protein was calculated from the [θ] data at 222 nm ([θ]222) based on the following equation53:
Quartz crystal microbalance-dissipation monitoring
Quartz crystal microbalance-dissipation (QCM-D) measurements were conducted using a QSense E4 instrument (Biolin Scientific AB, Stockholm, Sweden). A silica-coated AT-cut quartz crystal sensor chip with a fundamental frequency of 5 MHz (QSX 303, Biolin Scientific) was used to characterize protein adsorption onto silica surfaces. The temperature was set to 25 °C for all experiments, and the measurement operation was controlled by the QSoft 401 (v2.5.13.664) (Biolin Scientific) software package. Prior to each experiment, the sensor chips were sequentially rinsed with 1% (wt/vol) aqueous SDS solution, water, and ethanol, and then dried under a gentle stream of nitrogen gas, followed by treatment with oxygen plasma (PDC-002, Harrick Plasma, Ithaca, NY, USA) for 3 min. A peristaltic pump was used to inject liquid samples into the measurement chamber at a volumetric flow rate of 100 µL min−1. A stable baseline signal was first established in Tris buffer solution before a 100 µM BSA protein was introduced into the measurement chamber for 30 min, followed by a buffer washing step. The resonance frequency (ΔF) and energy dissipation (ΔD) shifts were recorded in real-time at multiple odd overtones, as previously described54, and the normalized data at the fifth overtone are reported.
The QCM-D technique was also employed to determine the passivation efficiency of BSA coatings against serum biofouling. For these experiments, 100 µM BSA solution was first added to a bare silica surface under continuous flow for 60 min, followed by a 40 min buffer washing step. Undiluted fetal bovine serum (FBS) was then introduced under continuous flow for 80 min, followed by a final buffer washing step for 30 min. The passivation efficiency percentage was calculated by comparing the absolute difference between the frequency shift due to BSA adsorption alone (after first buffer washing step) and the subsequent frequency shift due to FBS biofouling (after second buffer washing step), which is denoted as ΔFFBS-BSA. A control experiment without BSA coating was also conducted in order to measure the absolute frequency shift due to FBS biofouling alone (after buffer washing step), which is denoted as ΔFControl. For each BSA coating, the passivation efficiency percentage was calculated by the following equation:
Localized surface plasmon resonance
Ensemble-averaged localized surface plasmon resonance (LSPR) measurements were conducted in optical transmission mode using an Insplorion XNano instrument (Insplorion AB, Gothenburg, Sweden), as previously described55. A white light beam illuminating a circular area of ~4 mm2 is transmitted through a silica-coated sensor chip (Insplorion) consisting of well-separated gold nanodisk arrays (~8% surface coverage) deposited by hole-mask colloidal lithography56 on a transparent glass substrate. The nanodisks had an average height and diameter of 20 nm and 120 nm, respectively and the surface of the sensor chip was conformally coated with a silica film (~10 nm thick). The light was then passed through a quartz glass window and collected by a spectrophotometer. Prior to each experiment, the sensor chip was sequentially rinsed with 1% (wt/vol) aqueous SDS solution, water and ethanol, then dried under a gentle stream of nitrogen gas. The chip was then treated with oxygen plasma for 2.5 min. The sensor chip was then loaded into the measurement chamber and liquid samples were introduced by a peristaltic pump at a volumetric flow rate of 100 µL min−1. A stable baseline signal was established in Tris buffer solution before 100 µM BSA was introduced into the measurement chamber under continuous flow for 30 min, followed by a buffer washing step. The Insplorer software package (Insplorion AB) was used to record the LSPR extinction spectra with a time resolution of 1 Hz and the centroid (peak) position (λ) in the extinction spectrum at each time point was calculated by using a high-order polynomial fitting57. Thus, it was possible to determine the time-resolved Δλ shift due to protein adsorption and the time-derivative plot of the Δλ shift was also calculated using the OriginPro 2019b software package.
ATR-FTIR spectroscopy
Attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy experiments were conducted using a Bruker Vertex 70 FTIR spectrometer with a liquid-nitrogen-cooled mercury cadmium telluride (MCT) photodetector (Bruker, Baden-Württemberg, Germany) that was equipped with a MIRacle ATR accessory containing a three-reflection ZnSe ATR crystal (PIKE Technologies, Fitchburg, WI, USA). A 30 µL aliquot of 100 µM BSA solution was pipetted onto the ATR crystal to form a droplet, and an absorbance spectrum was immediately recorded to characterize the secondary structure of solution-phase BSA proteins. The BSA was then incubated with the ATR crystal surface at room temperature for 30 min, followed by a series of ten buffer washing steps to remove weakly adsorbed proteins. During each washing step, 30 µL of extra buffer solution (without protein) was added to the sample droplet, followed by aspiration of a 30 µL total volume from the droplet. After the washing steps were completed, then a second absorbance spectrum was recorded to characterize the secondary structure of BSA proteins in the adsorbed state. During measurements, the sample compartment was purged continuously with nitrogen gas to minimize ambient moisture. The OPUS 6.5 software package (Bruker) was used to collect the ATR-FTIR absorbance spectra at a 4 cm−1 resolution and averaged over 128 scans. All recorded spectra were normalized by subtracting background spectra from buffer and water vapor alone in order to remove water and ambient water vapor absorbance artefacts, respectively.
The relative contribution of different secondary structure elements was determined for all samples by curve fitting the amide I bands of the absorbance spectra (1720−1580 cm−1) to Gaussian components based on the peaks identified in the second derivative plot of the absorbance spectra and from literature references22,58. All curve fitting analyses were conducted using the Peak Analyzer function in the OriginPro 2019b software package. The absorbance spectrum obtained from experiment was assumed to be a linear combination of multiple constituent Gaussian peaks, each centered around a given wavenumber value and representing a protein secondary structure element. The secondary structure percentage values were determined by calculating the area under the curve-fitted peaks that had been assigned to each secondary structure element as a fraction of the total area under the fitted curve. Peaks centered at 1682–1678 cm−1 were assigned to β-turn structures, 1656–1654 cm−1 to α-helix, 1649–1648 cm−1 to random coil, 1639–1624 cm−1 to extended chains, and 1618–1612 cm−1 to intermolecular β-sheets.
SDS-PAGE and western blot
10% polyacrylamide resolving gels at pH 8.8 and 5% stacking gels at pH 6.8 with fifteen sample loading wells per gel were prepared before experiments. For sample preparation, normal human serum (NHS) was thawed and then immediately diluted to 1% (vol/vol) in Tris buffer, followed by incubation at 37 °C for 30 min. A 180 µL aliquot of diluted NHS was then mixed with 54 µL of 4× Laemmli buffer and 6 µL of 2-mercaptoethanol. The sample mixture was then heated to 95 °C for 5 min for complete denaturation of serum proteins, before cooling back to room temperature. A 10 µL aliquot of protein standards was loaded into the first lane of the polyacrylamide gel and the next two lanes were each loaded with 10 µL of the sample mixture; this sequence was repeated five times in total to fill all lanes within the gel. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was then conducted at 50 V for 15 min, followed by 100 V for 90 min in running buffer.
The protein bands in the gel were then transferred to a nitrocellulose membrane at 300 mA for 2 h in transfer buffer, and ice packs were used to keep the transfer buffer cool during the transfer process. After the transfer step, the membranes were cut into strips, each one containing the three lanes of protein standards (lane 1) and serum samples (lanes 2–3). Surface passivation (“blocking”) solutions were prepared using fatted or defatted BSA [3% (wt/vol) BSA in TBST; BSA 1 or 5] and each strip was incubated with 5 mL of the appropriate BSA blocking solution at room temperature for 1 h. The membranes were then incubated with complement C3b monoclonal antibody as the primary antibody (1:500 dilution in blocking solution) overnight. Each membrane was then washed with TBST for 10 min. After three washing steps, the membranes were incubated with HRP-conjugated goat anti-mouse IgG secondary antibody (1:2000 dilution in blocking solution) for 1 h, followed by four subsequent washing steps with TBST.
The membranes were then incubated with Clarity Max Western ECL substrate for 5 min in order to enhance chemiluminescence properties. All incubation and washing steps were conducted on a rocking platform. The protein bands were subsequently imaged using an Amersham Imager 600 instrument and its accompanying software (v0.9.8) (GE Healthcare, Chicago, IL, USA) with a 1 s exposure time. The Fiji/ImageJ software package59 was then used to quantify the intensity of the noise bands that were located near the 250, 75, and 50 kDa molecular weight markers. The digital image of the blots was first set to 8-bit greyscale, and then an intensity profile from each selected lane was plotted using the Analyze Gels function. The peaks from non-specific bands of interest were identified and a baseline was created by drawing a straight line between the two minima points on either side of each peak of interest. The Wand tool was then used to determine the area bounded by the peak and the baseline. These values were defined as the intensity values for each selected non-specific band.
Enzyme-linked immunosorbent assay
The MicroVue SC5b-9 Plus Enzyme Immunoassay kit (catalog no. A020; Quidel, San Diego, CA, USA) was used for enzyme-linked immunosorbent assay (ELISA) experiments. BSA-coated silica nanoparticle samples were prepared by mixing equal volumes of the appropriate 1 mg mL−1 BSA solution with 2 mg mL−1 silica nanoparticles in Tris buffer. The mixtures were then incubated at 37 °C for 2 h, followed by centrifugation at 16,000 × g for 30 min. The supernatants were removed and the nanoparticles were resuspended with fresh Tris buffer. Another round of centrifugation and resuspension was conducted to yield 1 mg mL−1 BSA-coated silica nanoparticle samples. A 10 µL aliquot of each BSA-coated silica nanoparticle sample was then mixed with a 40 µL aliquot of freshly thawed normal human serum (NHS). A 50 µL aliquot of NHS by itself was used as the negative control (lowest level of complement activation) and 10 µL of 1 mg mL−1 uncoated silica nanoparticles plus 40 µL NHS was used as the positive control (highest level of complement activation). All test samples were incubated at 37 °C for 30 min.
The assay was then performed using the 96-well plates that were provided in the ELISA kit according to the manufacturer’s instructions. All serum samples, including controls, were diluted 40-times by using the provided sample diluent solution before addition to the wells. Standard solutions were added without dilution. The absorbance values from each well were determined by an Infinite Pro 200 plate reader (Tecan, Männedorf, Switzerland). Standard curves were calculated by correlating the known concentrations of the standard solutions to the respective absorbance values. The SC5b-9 protein concentration in each sample was then computed. The degree of protection afforded by each BSA coating was calculated by treating the SC5b-9 protein concentrations from the negative and positive control samples as 100% and 0% protection, respectively.
Statistical analysis
Statistical analyses were conducted using the GraphPad Prism (v8.0.1) software package from GraphPad Software (La Jolla, CA, USA). One-way or two-way analysis of variance (ANOVA) with the appropriate multiple comparisons test and the unpaired t-test were used to compute the statistical significance of experimental data as appropriate. All statistical analysis involved two-tailed tests. Unpaired t-tests results are reported as P values while multiple comparisons test results are reported as multiplicity-adjusted P values. P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 indicate the levels of statistical significance. Additional information can be found in Supplementary Tables 6–14.
Data availability
All data supporting the findings of this study are available within this Article and its Supplementary Information files, and from the corresponding authors upon reasonable request.
References
Hucknall, A., Rangarajan, S. & Chilkoti, A. In pursuit of zero: polymer brushes that resist the adsorption of proteins. Adv. Mater. 21, 2441–2446 (2009).
Schöttler, S. et al. Protein adsorption is required for stealth effect of poly (ethylene glycol)-and poly (phosphoester)-coated nanocarriers. Nat. Nanotechnol. 11, 372 (2016).
Sapsford, K. E. et al. Functionalizing nanoparticles with biological molecules: developing chemistries that facilitate nanotechnology. Chem. Rev. 113, 1904–2074 (2013).
Wei, Q. et al. Protein interactions with polymer coatings and biomaterials. Angew. Chem. Int. Edit 53, 8004–8031 (2014).
Luo, N. et al. PEGylated graphene oxide elicits strong immunological responses despite surface passivation. Nat. Commun. 8, 1–10 (2017).
Sweryda-Krawiec, B., Devaraj, H., Jacob, G. & Hickman, J. J. A new interpretation of serum albumin surface passivation. Langmuir 20, 2054–2056 (2004).
Steinitz, M. Quantitation of the blocking effect of tween 20 and bovine serum albumin in ELISA microwells. Anal. Biochem. 282, 232–238 (2000).
Mahmood, T. & Yang, P.-C. Western blot: technique, theory, and trouble shooting. N. Am. J. Med. Sci. 4, 429 (2012).
Buchwalow, I. B. & Böcker, W. Immunohistochemistry: Basics and Methods 41–46 (Springer, Berlin Heidelberg, 2010).
Besecker, J., Cornell, K. A. & Hampikian, G. Dynamic passivation with BSA overcomes LTCC mediated inhibition of PCR. Sens. Actuators B 176, 118–123 (2013).
Convert, L. et al. Passivation of KMPR microfluidic channels with bovine serum albumin (BSA) for improved hemocompatibility characterized with metal-clad waveguides. Sens. Actuators B 173, 447–454 (2012).
Van Hong Nguyen, B.-J. L. Protein corona: a new approach for nanomedicine design. Int. J. Nanomed. 12, 3137 (2017).
del Río, J. S., Henry, O. Y., Jolly, P. & Ingber, D. E. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 14, 1143–1149 (2019).
Peng, Q. et al. Preformed albumin corona, a protective coating for nanoparticles based drug delivery system. Biomaterials 34, 8521–8530 (2013).
Mariam, J., Sivakami, S. & Dongre, P. M. Albumin corona on nanoparticles–a strategic approach in drug delivery. Drug Deliv. 23, 2668–2676 (2016).
Mannuzza, F. J. & Montalto, J. G. Is bovine albumin too complex to be just a commodity? BioProcess Int. 8, 42–49 (2010).
Xiao, Y. & Isaacs, S. N. Enzyme-linked immunosorbent assay (ELISA) and blocking with bovine serum albumin (BSA)—not all BSAs are alike. J. Immunol. Methods 384, 148–151 (2012).
Xia, X.-R., Monteiro-Riviere, N. A. & Riviere, J. E. An index for characterization of nanomaterials in biological systems. Nat. Nanotechnol. 5, 671–675 (2010).
Jeyachandran, Y., Mielczarski, J., Mielczarski, E. & Rai, B. Efficiency of blocking of non-specific interaction of different proteins by BSA adsorbed on hydrophobic and hydrophilic surfaces. J. Colloid Interface Sci. 341, 136–142 (2010).
Park, J. H. et al. Controlling adsorption and passivation properties of bovine serum albumin on silica surfaces by ionic strength modulation and cross-linking. Phys. Chem. Chem. Phys. 19, 8854–8865 (2017).
Park, J. H. et al. Temperature-induced denaturation of BSA protein molecules for improved surface passivation coatings. ACS Appl. Mater. Interfaces 10, 32047–32057 (2018).
Givens, B. E., Xu, Z., Fiegel, J. & Grassian, V. H. Bovine serum albumin adsorption on SiO2 and TiO2 nanoparticle surfaces at circumneutral and acidic pH: A tale of two nano-bio surface interactions. J. Colloid Interface Sci. 493, 334–341 (2017).
Dolatshahi-Pirouz, A. et al. Bovine serum albumin adsorption on nano-rough platinum surfaces studied by QCM-D. Colloids Surf. B 66, 53–59 (2008).
Satzer, P., Svec, F., Sekot, G. & Jungbauer, A. Protein adsorption onto nanoparticles induces conformational changes: particle size dependency, kinetics, and mechanisms. Eng. Life Sci. 16, 238–246 (2016).
Jachimska, B., Tokarczyk, K., Łapczyńska, M., Puciul-Malinowska, A. & Zapotoczny, S. Structure of bovine serum albumin adsorbed on silica investigated by quartz crystal microbalance. Colloids Surf. A 489, 163–172 (2016).
Cohn, E. J. et al. Preparation and properties of serum and plasma proteins. IV Is bovine albumin too complex to be just a commodity. J. Am. Chem. Soc. 68, 459–475 (1946).
Reid, A. F. Method of purifying albumin. US patent 2,705,230 (1955).
Porsche, J. D., Lesh, J. B. & Grossnickle, M. D. Recovery of serum albumin. US patent 2,765,299 (1956).
Schneider, W., Lefevre, H., Fiedler, H. & McCarty, L. J. An alternative method of large scale plasma fractionation for the isolation of serum albumin. Blut 30, 121–134 (1975).
Spector, A. A. Fatty acid binding to plasma albumin. J. Lipid Res. 16, 165–179 (1975).
Chen, R. F. Removal of fatty acids from serum albumin by charcoal treatment. J. Biol. Chem. 242, 173–181 (1967).
Dobson, C. M., Šali, A. & Karplus, M. Protein folding: a perspective from theory and experiment. Angew. Chem. Int. Edit 37, 868–893 (1998).
Militello, V. et al. Aggregation kinetics of bovine serum albumin studied by FTIR spectroscopy and light scattering. Biophys. Chem. 107, 175–187 (2004).
Jeyachandran, Y., Mielczarski, E., Rai, B. & Mielczarski, J. Quantitative and qualitative evaluation of adsorption/desorption of bovine serum albumin on hydrophilic and hydrophobic surfaces. Langmuir 25, 11614–11620 (2009).
Rabe, M., Verdes, D. & Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 162, 87–106 (2011).
Norde, W. Adsorption of proteins from solution at the solid-liquid interface. Adv. Colloid Interface Sci. 25, 267–340 (1986).
Frost, R. et al. Core–shell nanoplasmonic sensing for characterization of biocorona formation and nanoparticle surface interactions. ACS Sens. 1, 798–806 (2016).
Jackman, J. A., Ferhan, A. R. & Cho, N.-J. Nanoplasmonic sensors for biointerfacial science. Chem. Soc. Rev. 46, 3615–3660 (2017).
Jackman, J. A. et al. Indirect nanoplasmonic sensing platform for monitoring temperature-dependent protein adsorption. Anal. Chem. 89, 12976–12983 (2017).
Jackman, J. A. et al. Nanoplasmonic ruler to measure lipid vesicle deformation. Chem. Commun. 52, 76–79 (2016).
Zeng, H., Chittur, K. K. & Lacefield, W. R. Analysis of bovine serum albumin adsorption on calcium phosphate and titanium surfaces. Biomaterials 20, 377–384 (1999).
Roach, P., Farrar, D. & Perry, C. C. Surface tailoring for controlled protein adsorption: effect of topography at the nanometer scale and chemistry. J. Am. Chem. Soc. 128, 3939–3945 (2006).
Ricklin, D., Hajishengallis, G., Yang, K. & Lambris, J. D. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785 (2010).
Belling, J. N. et al. Stealth immune properties of graphene oxide enabled by surface-bound complement Factor H. ACS Nano 10, 10161–10172 (2016).
Chen, F. et al. Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo. Nat. Nanotechnol. 12, 387 (2017).
Curry, S., Mandelkow, H., Brick, P. & Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Mol. Biol 5, 827 (1998).
Curry, S. Plasma albumin as a fatty acid carrier. Adv. Mol. Cell Biol. 33, 29–46 (2003).
Karlsson, M., Ekeroth, J., Elwing, H. & Carlsson, U. Reduction of irreversible protein adsorption on solid surfaces by protein engineering for increased stability. J. Biol. Chem. 280, 25558–25564 (2005).
Ramsden, J. & Prenosil, J. Effect of ionic strength on protein adsorption kinetics. J. Phys. Chem. 98, 5376–5381 (1994).
Rial, R. et al. Structural and kinetic visualization of the protein corona on bioceramic nanoparticles. Langmuir 34, 2471–2480 (2018).
Galdino, F. E., Picco, A. S., Sforca, M. L., Cardoso, M. B. & Loh, W. Effect of particle functionalization and solution properties on the adsorption of bovine serum albumin and lysozyme onto silica nanoparticles. Colloids Surf. B 186, 110677 (2020).
Savitzky, A. & Golay, M. J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964).
Morrisett, J. D., David, J. S., Pownall, H. J. & Gotto, A. M. Jr. Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry 12, 1290–1299 (1973).
Cho, N.-J., Frank, C. W., Kasemo, B. & Höök, F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 5, 1096 (2010).
Jackman, J. A., Zhdanov, V. P. & Cho, N.-J. Nanoplasmonic biosensing for soft matter adsorption: kinetics of lipid vesicle attachment and shape deformation. Langmuir 30, 9494–9503 (2014).
Fredriksson, H. et al. Hole–mask colloidal lithography. Adv. Mater. 19, 4297–4302 (2007).
Dahlin, A. B., Tegenfeldt, J. O. & Höök, F. Improving the instrumental resolution of sensors based on localized surface plasmon resonance. Anal. Chem. 78, 4416–4423 (2006).
Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 1767, 1073–1101 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676 (2012).
Acknowledgements
The authors thank Mr. Tun Naw Sut for technical assistance with QCM-D experiments. This work was supported by the National Research Foundation of Singapore through a Competitive Research Programme grant (NRF-CRP10-2012-07) and a Proof-of-Concept grant (NRF2015NRF-POC0001-19), and by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1C1C1004385).
Author information
Authors and Affiliations
Contributions
G.J.M., A.R.F., J.A.J., and N.-J.C. planned the studies. G.J.M. and A.R.F. conducted experiments. G.J.M., A.R.F., J.A.J., and N.-J.C. interpreted the results. J.A.J. and N.-J.C. obtained funding. G.J.M., A.R.F., and J.A.J. wrote the first draft of the manuscript. All authors reviewed, edited and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, G.J., Ferhan, A.R., Jackman, J.A. et al. Conformational flexibility of fatty acid-free bovine serum albumin proteins enables superior antifouling coatings. Commun Mater 1, 45 (2020). https://doi.org/10.1038/s43246-020-0047-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43246-020-0047-9
This article is cited by
-
Antifouling strategies for electrochemical sensing in complex biological media
Microchimica Acta (2024)
-
Microfluidic Capture Device for Simple, Cell Surface Marker-Based Quantification of Senescent CD8+ T Cells
BioChip Journal (2024)
-
Mixed Matrix Cellulose Acetate Membrane Incorporated with Poly(amidoamine) Dendrimer Functionalized Multiwalled Carbon Nano Tubes for Effective Anionic Dyes Rejection
Journal of Inorganic and Organometallic Polymers and Materials (2023)
-
Difference in the binding mechanism of distinct antimony forms in bovine serum albumin
BioMetals (2021)