Abstract
Among the greatest hurdles in clinical management of prostate cancer (PCa) are the progression to lethal castration-resistant prostate cancer (CRPC) and the lack of suitable targeted therapies for advanced disease. Here we identify Gremlin1 as a ligand for fibroblast growth factor receptor 1 (FGFR1), which promotes lineage plasticity and drives castration resistance. Importantly, we generate a specific anti-Gremlin1 therapeutic antibody and demonstrate synergistic effect with androgen deprivation therapy (ADT) in CRPC. GREM1 transcription is suppressed by androgen receptor (AR) and released following ADT. We show that Gremlin1 binds to FGFR1 and activates downstream MAPK signaling. Gremlin1 interacts with FGFR1 differently to its canonical ligand FGF1, as revealed through protein structure docking and mutagenesis experiments. Altogether, our data indicate Gremlin1 as a promising candidate therapeutic target for CRPC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Anti-human Gremlin1 antibody and anti-mouse Gremlin1 antibody were generated in this study. Deep-sequencing (ChIP–seq and RNA–seq) data that support the findings of this study have been deposited in the National Omics Data Encyclopedia, with accession number OEP001758. Previously published microarray data that were reanalyzed here are available at www.oncomine.org. The human PCa transcriptome data that support the findings of this study are available at www.cbioportal.com. The data from Beltran et. al. generated for this study are available through dbGaP phs000909 and can be accessed by agreement.
Source data for Figs. 1–7 and Extended Data Figs. 1–10 have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Scher, H. I. et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012).
Ryan, C. J. et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013).
Tran, C. et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009).
Davies, A. H., Beltran, H. & Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 15, 271–286 (2018).
Hsu, D. R., Economides, A. N., Wang, X., Eimon, P. M. & Harland, R. M. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1, 673–683 (1998).
Verheyden, J. M. & Sun, X. An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature 454, 638–641 (2008).
Benazet, J. D. et al. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science 323, 1050–1053 (2009).
Costello, C. M., Cahill, E., Martin, F., Gaine, S. & McLoughlin, P. Role of gremlin in the lung: development and disease. Am. J. Respir. Cell Mol. Biol. 42, 517–523 (2010).
Boers, W. et al. Transcriptional profiling reveals novel markers of liver fibrogenesis: Gremlin and insulin-like growth factor-binding proteins. J. Biol. Chem. 281, 16289–16295 (2006).
Yan, K. et al. Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 28, 1085–1100 (2014).
Davis, H. et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 21, 62–70 (2015).
Myllarniemi, M. et al. Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 177, 321–329 (2008).
Carter, P. J. & Lazar, G. A. Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nat. Rev. Drug Discov. 17, 197–223 (2018).
Best, C. J. et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin. Cancer Res. 11, 6823–6834 (2005).
Yu, Y. P. et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 22, 2790–2799 (2004).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Bluemn, E. G. et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489 (2017).
Su, W. et al. The Polycomb Repressor Complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell 36, 139–155 (2019).
Abida, W. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019).
Bennett, J. T. et al. Mosaic activating mutations in FGFR1 cause encephalocraniocutaneous lipomatosis. Am. J. Hum. Genet. 98, 579–587 (2016).
Plotnikov, A. N., Hubbard, S. R., Schlessinger, J. & Mohammadi, M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101, 413–424 (2000).
Hou, J. et al. Substitution of putative half-cystine residues in heparin-binding fibroblast growth factor receptors. Loss of binding activity in both two and three loop isoforms. J. Biol. Chem. 267, 17804–17808 (1992).
Koika, V. et al. Comparative functional analysis of two fibroblast growth factor receptor 1 (FGFR1) mutations affecting the same residue (R254W and R254Q) in isolated hypogonadotropic hypogonadism (IHH). Gene 516, 146–151 (2013).
Raivio, T. et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 94, 4380–4390 (2009).
Kisonaite, M., Wang, X. & Hyvonen, M. Structure of Gremlin-1 and analysis of its interaction with BMP-2. Biochem. J 473, 1593–1604 (2016).
Beenken, A., Eliseenkova, A. V., Ibrahimi, O. A., Olsen, S. K. & Mohammadi, M. Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J. Biol. Chem. 287, 3067–3078 (2012).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Lunardi, A. et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat. Genet. 45, 747–755 (2013).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Li, Z. G. et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J. Clin. Invest. 118, 2697–2710 (2008).
Cappellen, D. et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 23, 18–20 (1999).
Deng, N. et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 61, 673–684 (2012).
Weiss, J. et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2, 62ra93 (2010).
Formisano, L. et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 10, 1373 (2019).
Ku, S. Y. et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017).
Mu, P. et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017).
Zou, M. et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 7, 736–749 (2017).
Labrecque, M. P. et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest. 129, 4492–4505 (2019).
Bishop, J. L. et al. The master neural rtanscription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 7, 54–71 (2017).
Svensson, C. et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 42, 999–1015 (2014).
Worthley, D. L. et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell 160, 269–284 (2015).
Quante, M. et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19, 257–272 (2011).
Rowan, S. C. et al. Gremlin 1 depletion in vivo causes severe enteropathy and bone marrow failure. J. Pathol. 251, 117–122 (2020).
Hieronymus, H. et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330 (2006).
Zhu, X. et al. Ubiquitination of inositol-requiring enzyme 1 (IRE1) by the E3 ligase CHIP mediates the IRE1/TRAF2/JNK pathway. J. Biol. Chem. 289, 30567–30577 (2014).
Zhou, Z. et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology 147, 1043–1054 (2014).
Yan, Y., Zhang, D., Zhou, P., Li, B. & Huang, S. Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 45, W365–W373 (2017).
Drost, J. et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016).
Zhang, K. et al. WNT/beta-catenin directs self-renewal symmetric cell division of hTERT(high) prostate cancer stem cells. Cancer Res. 77, 2534–2547 (2017).
Acknowledgements
The study was supported by funds to H.H.Z. from the National Key R&D Program of China (no. 2017YFA0102900), the National Natural Science Foundation of China (nos. NSFC32022021, and NSFC81972404), the Program of Shanghai Academic/Technology Research Leader (no. 21XD1422300), the Shanghai Municipal Education Commission–Gaofeng Clinical Medicine Grant Support (no. 20181706) and the Innovative research team of high-level local universities in Shanghai; and by funds to W.-Q.G. from NSFC (nos. 81630073 and 81872406), 111 project (no. B21024), the Science and Technology Commission of Shanghai Municipality (nos. 20JC1417600 and 21JC1404100), Peak Disciplines (Type VI) of Institutes of Higher Learning in Shanghai, the Shanghai Jiao Tong University STAR and Scientific and Technological Innovation Funds and the KC Wong foundation. Funding for the generation and production of antibodies for Gremlin1 were provided by Mabspace Biosciences.
Patient-derived organoid lines BM1, BM61, ST88 and ST60 were kindly provided by Prof. Dong Gao (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China). Patient-derived PDX MDA PCa 118b was generously shared by Prof. Nora M. Navone (Department of Genitourinary Medical Oncology) and the David H. Koch Center for Applied Research of Genitourinary Cancers, the University of Texas MD Anderson Cancer Center, Houston, Texas.
Author information
Authors and Affiliations
Contributions
H.H.Z. and W.-Q.G. conceived the study. X.Q. contributed to antibody-related study design. C.C. and J.W. performed the experiments. L.F., Y.G., Z.X., B.D. and W.X. collected clinical patient samples. K.Z. provided support for in vivo experiments. D.W., N.J., Z.J., L.F. and H.H.Z. helped with cell culture experiments. Y.H. and Y.G. participated in immunostaining and imaging. P.X., M.Z., K.L. and P.Z. assisted with animal experiments. J.Z. and P.Z. supported data analysis. C.L. provided assistance with ChIP experiments. B.D. and W.X. provided clinical patient samples, patient-derived cell lines and histological assessment. S.C., Z.S., D.S., X.Y., H.L. and X.Q. provided support for antibody generation, engineering, production and characterization. H.H.Z., W.-Q.G., C.C. and J.W. interpreted the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
X.Q., Z.S., D.S., X.Y. and H.L. are employees and shareholders of Mabspace Biosciences (Suzhou) Co., Ltd.
Peer review
Peer review information
Nature Cancer thanks Leigh Ellis, John Isaacs and Ping Mu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
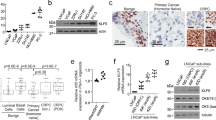
Extended Data Fig. 1 Gremlin1 is upregulated in CRPCs and associated with a poor disease outcome.
(a) Analysis of Oncomine PCa dataset (Yu dataset (GSE6919), n = 64 tumor samples in primary site; n = 24 CRPC samples) demonstrates that the GREM1 expression level is significantly upregulated in castration resistant PCa. (b) The GREM1 gene transcription is significantly upregulated in DNPC (data were obtained from the Beltran 2016 dataset (dbGaP phs000909), n = 25 ARPC samples, n = 11 DNPC samples, n = 13 NEPC samples). (c) A significant shorter disease/progression-free survival time in patients carrying GREM1 gene copy number amplifications compared to patients without GREM1 DNA alterations (left panel) or in patients with higher GREM1 mRNA expression (right panel). (d) High expression of Gremlin1 has a positive correlation with poor overall survival in SU2C 2019 dataset (dbGaP phs000915). (e-f) GREM1 mRNA exhibits a negative correlation with AR score in the SU2C 2019 dataset, total n = 266 tumor samples, n = 133 tumor samples in the group of high AR score, n = 133 tumor samples in the group of low AR score. The AR score here was acquired from the cBioPortal database which contains 30 AR responsive genes described previously. FPKM, fragments per kilobase million. (g) The treatment time on AR targeted therapy in patient groups with high or low expression level of Gremlin1 in the SU2C 2019 dataset (dbGaP phs000915), n = 28 tumor samples in the group of GREM1 high, n = 28 tumor samples in the group of GREM1 low. (Two-tailed Student’ s t test was used for the statistical analysis. Log-rank (Mantel-Cox) test was used for the survival analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 2 Transcriptional suppression of AR on GREM1 depends on a high androgen concentration.
(a-i) Immunoblotting and q-PCR analysis of GREM1 expression, and GREM1 promoter-driven luciferase assay in AR overexpressed LNCaP (a, b, g) or LAPC4 cells (c, d, h) and AR knock-out LNCaP cells (e, f, i) (b, d, n = 4 independently treated cell cultures, f, n = 3 independently treated cell cultures, g, h, i, n = 3 independently transfected replicates). (j, k) The AR gene transcription is upregulated in LNCaP-R and VCAP cells treated with charcoal/dextran-stripped FBS for 3 weeks (VCAP-R) (n = 3 independently treated cell cultures). (l, m) LNCaP or LNCaP-R cells and VCAP or VCAP-R cells were treated with 0, 0.01, 0.1, 1, or 10 nM DHT for 24 hrs. GREM1, KLK3 and OPRK1 mRNA are measured by q-PCR analysis (n = 3 independently treated cell cultures). (n) 0, 0.1 or 10 nM DHT were added to LNCaP cells for 4 hr. Binding of AR to the ARE of GREM1 or KLK3 gene is measured by ChIP q-PCR. (o) LNCaP-R cells were treated with or without 10 nM DHT. The enrichment of AR to GREM1 and KLK3 gene is measured by ChIP q-PCR (n = 3 independently treated cell cultures). (Two-tailed Student’ s t test was used for the statistical analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 3 Gremlin1 promotes PCa cell proliferation, castration resistance and tumor growth in vivo.
(a) Immunoblotting confirms the efficiency of GREM1 knockdown or GREM1 overexpression in PC3 cells. (b, c) GREM1 knockdown leads to a suppression of sphere formation capacity (n = 3 biological replicates) (b) and cell proliferation (n = 3 independently treated cell cultures) (c) in PC3 cells, while GREM1 overexpression or addition of exogenous Gremlin1 protein display a promoting effect. (d) Knockdown of GREM1 increases cell apoptosis in PC3 cells (n = 3 independently treated cell cultures). (e) GREM1 knockdown represses PC3 xenografts growth in vivo. n = 8 mice. (f) GREM1 knockdown increases cell apoptosis of PC3 xenografts in vivo characterized by the level of cleaved caspase3. (g) GREM1 overexpression promotes PC3 xenografts forming incidence and tumor growth in vivo. Scale bars = 1 cm. n = 6 mice in the xenograft experiments with 1×106 and 1×104 cells. n = 4 mice in the 1×104 cell xenograft assay. (h) Immunoblotting confirms Gremlin1 overexpression in LAPC4 cells transfected with GREM1 lentivirus. (i) Gremlin1 enhances sphere forming of LAPC4 cells (n = 3 biological replicates). (j) Gremlin1 promotes the growth of LAPC cells under the ADT treatment (n = 3 independently treated cell cultures). (k) GREM1 overexpression prevents cell death upon enzalutamide treatment characterized by a decreased Annexin V/DAPI staining (n = 3 independently treated cell cultures). Immunoblotting was repeated at least 3 times and representative images were shown. ADT: treated with enzalutamide at 10 μg/ml. (Two-tailed Student’ s t test was used for the statistical analysis. Two-way ANOVA analysis were used for the measurement of tumor volume. Data are presented as means ± s.e.m.).
Extended Data Fig. 4 Gremlin1 induces a decrease in AR signaling and alterations in expression of cell lineage genes in PCa cells.
(a, b) q-PCR assessment of transcription of AR signaling genes, stem cell-related genes, basal, luminal, and neuroendocrine lineage makers in control and GREM1 overexpressing LNCaP (a) or LAPC4 (b) cells (n = 3 independently treated cell cultures). (c, d) ARE-luciferase reporter assay demonstrates an AR signaling decrease upon GREM1 overexpression in LNCaP (c) (n = 3 independently transfected replicates) or LAPC4 (d) cells (n = 4 independently transfected replicates). (e, f) Gene set enrichment analysis shows a downregulation of AR signaling and luminal genes, and an enrichment of basal cell signature and stemness genes in Gremlin1 overexpressed LNCaP cells (e) and Hi-Myc mouse PCa derived organoids (f) (n = 3 independent samples). (g) ChIP-seq analysis indicate a reduced AR chromatin binding intensity at AR target genes.
Extended Data Fig. 5 Gremlin1 promotes PCa in a FGFR1/MAPK-dependent manner.
(a) Gremlin1 treatment leads to an activation of FGFR1/MEK/ERK signaling pathway in a dose-dependent way. FGF1 and FGF2 are used as positive controls. Concentration (ng/ml) of Gremlin1, FGF1 and FGF2 are shown in the figure. (b) FGFR1/MEK/ERK signaling pathway is activated in the GREM1 overexpressed PC3 and LNCaP cells. GREM1 overexpression also promotes FGFR2 and AKT phosphorylation. (c) Immunoblotting shows the efficiency of FGFR1 knockout in LNCaP cells. (d, e) FGFR1 knockout significantly attenuates the positive effects of Gremlin1 (100 ng/ml) on LNCaP-R cell proliferation (n = 3 independently treated cell cultures) and sphere formation (n = 3 biological replicates). (f, g) The promoting effects of Gremlin1 protein (100 ng/ml) on LNCaP-R cells proliferation (f, n = 6 independently treated cell cultures) and sphere forming (g, n = 4 biological replicates) are abrogated by FGFR1 or MEK inhibitors but are not affected by the addition of BMP4 or BMP6. BGJ398, FGFR1 inhibitor; Trametinib, MEK inhibitor. (h, i) FGFR1 activation promotes LNCaP cell growth in ADT condition (f, n = 4 independently treated cell cultures) or upon AR knock-out (g, n = 5 independently treated cell cultures). LNCaP or LNCaP sgAR cells were transfected with plasmids expressing FGFR1 N546K or FGFR1 R656P constitutively activated mutation, or treated with FGFR1 inhibitor BGJ398. (j) Addition of Gremlin1 protein enhances the cell proliferation in LNCaP AR knock-out cells (n = 5 independently treated cell cultures). Immunoblotting was repeated at least 3 times and representative images were shown. (Two-tailed Student’ s t test was used for the statistical analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 6 Gremlin1 binds to FGFR1 in a way different from FGF1/FGFR1 interaction.
(a) The specific binding of Gremlin1 to immobilized FGFR1 is competed by soluble FGFR1 in a dose-dependent manner (n = 2 technical replicates, experiments have been repeated twice). (b) Soluble FGFR1 (200 ng/ml) competitively inhibits the activation of FGFR1/MEK/ERK signaling by Gremlin1 (100 ng/ml) in PC3. The experiments were repeated at least 3 times with similar results. (c) Schematics of FGFR1 mutations. (d) FGFR1-C176G or FGFR1-R248Q mutation abolishes co-immunoprecipitation of FGF1 and FGFR1(left panel), but do not influence the forming of protein complex between Gremlin1 and FGFR1(right panel). The experiments were repeated at least 3 times with similar results. (e-h) The binding between Gremlin1 and FGFR1 is not affected by addition of FGF1, and vice versa, which are revealed by Fortebio (e), co-immunostaining (f), pull-down (g), and immunoblotting (h) assays. Scale bars = 10 μm. The Fortebio experiment was repeated at least twice with similar results. The pull-down and immunoblotting assay were repeated at least 3 times with similar results. The immunostaining experiment was repeated at least twice. Multiple fields of cell culture slides were examined during each repeat and representative images were shown. (i) mRNA levels of GREM1 and FGFs in CRPCs (data were obtained from the Beltran 2016 dataset (dbGaP, phs000909), n = 49 tumor samples). (j) Correlation analysis of GREM1 and FGFs expression in CRPCs (data were obtained from the Beltran 2016 dataset (dbGaP, phs000909), n = 49 tumor samples). (k) The concentration of Gremlin1 and FGF1 in prostatic fluid of human PCa patients is analyzed by the enzyme-linked immunosorbent assay. n = 18 prostatic fluid samples in Gremlin1 ELISA assay; n = 23 prostatic fluid samples in FGF1 ELISA assay. (Two-tailed Student’ s t test was used for the statistical analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 7 Gremlin1 antibody treatment in mice does not cause major side effects in vivo.
(a) Binding specificity of anti-murine Gremlin1 antibody to Gremlin1 is validated by the enzyme-linked immunosorbent assay (n = 2 technical replicates, experiments have been repeated twice). Ab is anti-Gremlin1 in this figure. (b) Gremlin was mainly expressed by the tumorous epithelial cells in castrated Pbsn-Cre4; Ptenfl/fl; Trp53fl/fl PCa. Scale bars = 20μm. The immunostaining experiment was repeated at least twice. The representative images were presented. (c) Grem1 was most highly expressed in the prostate cancer tissue compared to other organs from Pbsn-Cre4; Ptenfl/fl; Trp53fl/fl mice (n = 3 mice). (d, e) Gremlin1 antibody treatment does not induce major side effects when administered systemically to mice (10 mg/kg three times a week). No obvious alterations are detected in peripheral blood cell counts (d, IgG: n = 5 mice; Ab: n = 7 mice) or major organs (e) in mice received the antibody treatment. Scale bars: left panel = 2 mm; right panel = 100 μm. Ab: anti-murine Gremlin1. (Two-tailed Student’ s t test was used for the statistical analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 8 Gremlin1 antibody treatment suppresses stem cell-related gene expression, whereas upregulates AR signaling and luminal signature genes in murine Pbsn-Cre4; Ptenfl/fl; Trp53fl/fl PCa.
(a-e) q-PCR (a) and Gene set enrichment analysis (b-e) on stem cell-related gene, AR signaling, luminal, basal, and neuroendocrine signatures on anti-Gremlin1 antibody-treated murine Pbsn-Cre4; Ptenfl/fl; Trp53fl/fl prostate cancer tissue (n = 3 mice). FDR, false discovery rate. (Two-tailed Student’ s t test was used for the statistical analysis. Data are presented as means ± s.e.m.).
Extended Data Fig. 9 Antitumor effects of the Gremlin1 antibody are acted through the FGFR1 inhibition.
(a) Binding specificity of the anti-human Gremlin1 to Gremlin1 is validated by the enzyme-linked immunosorbent assay. Ab is anti-human Gremlin1 in this figure. (b) The antibody against Gremlin1 (100 ng/ml) facilitates the inhibition of in vitro cell proliferation by enzalutamide (10 μg/ml) (n = 3 independently treated cell cultures). (c) Anti-Gremlin1 treatment suppresses the sphere formation ability of LNCaP-R cells (n = 3 biological replicates). (d) Annexin-V/DAPI staining demonstrates that anti-Gremlin1 antibody displays a synergistic effect with enzalutamide in inducing cell death (n = 2 independently treated cell cultures, experiments have been repeated at least 3 times). (e) The activation of FGFR1/MEK/ERK signaling pathway is suppressed by the Gremlin1 antibody in LNCaP-R cells. The experiment was repeated at least 3 times with similar results. (f) Immunoblotting confirms the efficiency of BMPR2 knockout in LNCaP-R cells. The experiment was repeated at least 3 times with similar results. (g) BMPR2 knockout shows no significant influence to the inhibitory effect of Gremlin1 antibody (10 μg/ml) on LNCaP-R cell proliferation (n = 3 independently treated cell cultures) and sphere formation (n = 3 biological replicates). Ab: anti-Gremlin1. ADT: treated with enzalutamide at 10 μg/ml. (h) Anti-Gremlin1 antibody (10 μg/ml) suppresses PCa PDO formation in a serial organoid forming assay (n = 3 biological replicates). (i) Expression levels of FGFR1 in four patient-derived xenograft lines are assessed by immunoblotting. The experiment was repeated at least 3 times with similar results. (j) FGFR inhibitor BGJ398 decreases organoid forming capacity of BM1 and BM61 PDX lines. No additive effect is detected between BGJ398 (1 μM) and Gremlin1 antibody (10 μg/ml) treatment (n = 3 biological replicates). Scale bars= 50 μm. FGFRi, FGFR1 inhibitor BGJ398. (Two-tailed Student’ s t test was used for the statistical analysis and the tumor size at the end timepoint was analyzed. Data are presented as means ± s.e.m.).
Extended Data Fig. 10 Validation of the anti-human Gremlin1 antibody for various applications and gating strategy for flow cytometry in this study.
(a-c) Antibody specificity of the anti-human Gremlin1 antibody (Mabspace Biosciences) in immunoprecipitation, immunoblotting, IHC and IF was validated using GREM1 overexpressing, GREM1 knockout and respective control LNCaP cells. IHC Scale bars = 50 μm, IF Scale bars = 20 μm. The immunostaining experiment was repeated at least twice and representative images were presented. The immunoblotting experiment was repeated at least 3 times with similar results. (d) Mass spectrum of immunoprecipitants with the anti-human Gremlin1 antibody (Mabspace Biosciences) in LNCaP cells shows an enrichment of the Gremlin1 protein. The experiment was repeated at least 3 times with similar results. (e) Gating strategy for the flow cytometric examination of apoptosis in this study. The frequency of the cells in the apoptotic stage were calculated based on APC + /DAPI−.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 1
Unprocessed immunoblots.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 2
Unprocessed immunoblots.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 3
Unprocessed immunoblots.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 4
Unprocessed immunoblots.
Source Data Fig. 5
Unprocessed immunoblots.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 6
Unprocessed immunoblots.
Source Data Fig. 7
Statistical Source Data.
Source Data Fig. 7
Unprocessed immunoblots.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 2
Unprocessed immunoblots.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 3
Unprocessed immunoblots.
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 6
Unprocessed immunoblots.
Source Data Extended Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 8
Statistical Source Data.
Source Data Extended Data Fig. 9
Statistical Source Data.
Source Data Extended Data Fig. 9
Unprocessed immunoblots.
Source Data Extended Data Fig. 10
Unprocessed immunoblots.
Rights and permissions
About this article
Cite this article
Cheng, C., Wang, J., Xu, P. et al. Gremlin1 is a therapeutically targetable FGFR1 ligand that regulates lineage plasticity and castration resistance in prostate cancer. Nat Cancer 3, 565–580 (2022). https://doi.org/10.1038/s43018-022-00380-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-022-00380-3
This article is cited by
-
FGFR-targeted therapeutics: clinical activity, mechanisms of resistance and new directions
Nature Reviews Clinical Oncology (2024)
-
OTUD4 promotes the progression of glioblastoma by deubiquitinating CDK1 and activating MAPK signaling pathway
Cell Death & Disease (2024)
-
Label-free biosensing with singular-phase-enhanced lateral position shift based on atomically thin plasmonic nanomaterials
Light: Science & Applications (2024)
-
Molecular panorama of therapy resistance in prostate cancer: a pre-clinical and bioinformatics analysis for clinical translation
Cancer and Metastasis Reviews (2024)
-
Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome
Genome Medicine (2023)