Abstract
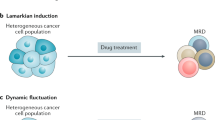
Stochastic transition of cancer cells between drug-sensitive and drug-tolerant persister phenotypes has been proposed to play a key role in non-genetic resistance to therapy. Yet, we show here that cancer cells actually possess a highly stable inherited chance to persist (CTP) during therapy. This CTP is non-stochastic, determined pre-treatment and has a unimodal distribution ranging from 0 to almost 100%. Notably, CTP is drug specific. We found that differential serine/threonine phosphorylation of the insulin receptor substrate 1 (IRS1) protein determines the CTP of lung and of head and neck cancer cells under epidermal growth factor receptor inhibition, both in vitro and in vivo. Indeed, the first-in-class IRS1 inhibitor NT219 was highly synergistic with anti-epidermal growth factor receptor therapy across multiple in vitro and in vivo models. Elucidation of drug-specific mechanisms that determine the degree and stability of cellular CTP may establish a framework for the elimination of cancer persisters, using new rationally designed drug combinations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Mass spectrometry phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE61 partner repository with the dataset identifiers PXD026824, PXD026842, PXD026844, PXD026857 and PXD026805. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE61 partner repository with the dataset identifier PXD026834. RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession codes GSE178978 and GSE179240. Source data are provided with this paper.
Code availability
Code for Extended Data Fig. 10 is found at https://doi.org/10.5281/zenodo.5036166.
References
Gupta, P. B. et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011).
Chaffer, C. L. et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl Acad. Sci. USA 108, 7950–7955 (2011).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Knoechel, B. et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat. Genet. 46, 364–370 (2014).
Lee, M.-C. W. et al. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc. Natl Acad. Sci. USA 111, E4726–E4735 (2014).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Fallahi-Sichani, M., Honarnejad, S., Heiser, L. M., Gray, J. W. & Sorger, P. K. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat. Chem. Biol. 9, 708–714 (2013).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Bigger, J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944).
Hata, A. N. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. https://doi.org/10.1038/nm.4040 (2016).
Liao, B.-C., Lin, C.-C. & Yang, J. C.-H. Second and third-generation epidermal growth factor receptor tyrosine kinase inhibitors in advanced nonsmall cell lung cancer. Curr. Opin. Oncol. 27, 94–101 (2015).
Pao, W. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73 (2005).
Fridman, O., Goldberg, A., Ronin, I., Shoresh, N. & Balaban, N. Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014).
Pearl Mizrahi, S., Gefen, O., Simon, I. & Balaban, N. Q. Persistence to anti-cancer treatments in the stationary to proliferating transition. Cell Cycle https://doi.org/10.1080/15384101.2016.1248006 (2016).
Hangauer, M. J. et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017).
Terai, H. et al. ER stress signaling promotes the survival of cancer ‘persister cells’ tolerant to EGFR tyrosine kinase inhibitors. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-17-1904 (2017).
Tetsu, O., Phuchareon, J., Eisele, D. W., Hangauer, M. J. & McCormick, F. AKT inactivation causes persistent drug tolerance to EGFR inhibitors. Pharmacol. Res. 102, 132–137 (2015).
Raha, D. et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-13-3456 (2014).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature https://doi.org/10.1038/nature22794 (2017).
Kokubo, Y. et al. Reduction of PTEN protein and loss of epidermal growth factor receptor gene mutation in lung cancer with natural resistance to gefitinib (IRESSA). Br. J. Cancer 92, 1711–1719 (2005).
Raoof, S. et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene https://doi.org/10.1038/s41388-019-0887-2 (2019).
Shah, K. N. et al. Aurora kinase A drives the evolution of resistance to third generation EGFR inhibitors in lung cancer. Nat. Med. 25, 111–118 (2019).
Wakata, K. et al. A favourable prognostic marker for EGFR mutant non-small cell lung cancer: immunohistochemical analysis of MUC5B. BMJ Open 5, e008366 (2015).
Corominas-Faja, B. et al. Stem cell-like ALDH(bright) cellular states in EGFR-mutant non-small cell lung cancer: a novel mechanism of acquired resistance to erlotinib targetable with the natural polyphenol silibinin. Cell Cycle 12, 3390–3404 (2013).
Shaw, L. M. The insulin receptor substrate (IRS) proteins: at the intersection of metabolism and cancer. Cell Cycle 10, 1750–1756 (2011).
Copps, K. D. & White, M. F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 55, 2565–2582 (2012).
Li, L. et al. Acquisition of EGFR TKI resistance and EMT phenotype is linked with activation of IGF1R/NF-κB pathway in EGFR-mutant NSCLC. Oncotarget 8, 92240–92253 (2017).
Cassell, A. & Grandis, J. R. Investigational EGFR-targeted therapies in HNSCC. Expert Opin. Investig. Drugs 19, 709–722 (2010).
Grandis, J. R. & Tweardy, D. J. Elevated levels of transforming growth factor α and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 53, 3579–3584 (1993).
Ozanne, B., Richards, C. S., Hendler, F., Burns, D. & Gusterson, B. Over-expression of the EGF receptor is a hallmark of squamous cell carcinomas. J. Pathol. 149, 9–14 (1986).
Kanety, H., Feinstein, R., Papa, M. Z., Hemi, R. & Karasik, A. Tumor necrosis factor α-induced phosphorylation of insulin receptor substrate-1 (IRS-1). Possible mechanism for suppression of insulin-stimulated tyrosine phosphorylation of IRS-1. J. Biol. Chem. 270, 23780–23784 (1995).
El-Ami, T. et al. A novel inhibitor of the insulin/IGF signaling pathway protects from age-onset, neurodegeneration-linked proteotoxicity. Aging Cell 13, 165–174 (2014).
Ibuki, N. et al. The tyrphostin NT157 suppresses insulin receptor substrates and augments therapeutic response of prostate cancer. Mol. Cancer Ther. 13, 2827–2839 (2014).
Reuveni, H. et al. Therapeutic destruction of insulin receptor substrates for cancer treatment. Cancer Res. 73, 4383–4394 (2013).
Ramirez, M. et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat. Commun. 7, 10690 (2016).
Russo, M. et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science https://doi.org/10.1126/science.aav4474 (2019).
Banelli, B. et al. The histone demethylase KDM5A is a key factor for the resistance to temozolomide in glioblastoma. Cell Cycle 14, 3418–3429 (2015).
Murakami, A. et al. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS ONE 9, e86459 (2014).
Vinogradova, M. et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12, 531–538 (2016).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Guler, G. D. et al. Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell https://doi.org/10.1016/j.ccell.2017.07.002 (2017).
Shaffer, S. M. et al. Memory sequencing reveals heritable single-cell gene expression programs associated with distinct cellular behaviors. Cell 182, 947–959.e17 (2020).
Spencer, S. L., Gaudet, S., Albeck, J. G., Burke, J. M. & Sorger, P. K. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009).
Faber, A. C. et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 1, 352–365 (2011).
Faber, A. C. et al. Epithelial-to-mesenchymal transition antagonizes response to targeted therapies in lung cancer by suppressing BIM. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-17-1577 (2017).
Song, K.-A. et al. Epithelial-to-mesenchymal transition antagonizes response to targeted therapies in lung cancer by suppressing BIM. Clin. Cancer Res. 24, 197–208 (2018).
Rehman, S. K. et al. Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell 184, 226–242 (2021).
Zhang, Y. Persisters, persistent infections and the yin–yang model. Emerg. Microbes Infect. 3, e3 (2014).
Raj, A. & Oudenaarden, Avan Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Barria, A., Muller, D., Derkach, V., Griffith, L. C. & Soderling, T. R. Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science 276, 2042–2045 (1997).
Lee, H. K., Barbarosie, M., Kameyama, K., Bear, M. F. & Huganir, R. L. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature 405, 955–959 (2000).
Sweatt, J. D. Memory mechanisms: the yin and yang of protein phosphorylation. Curr. Biol. 11, R391–R394 (2001).
Langlais, P. et al. Global IRS-1 phosphorylation analysis in insulin resistance. Diabetologia 54, 2878–2889 (2011).
Yarchoan, M. et al. Abnormal serine phosphorylation of insulin receptor substrate 1 is associated with tau pathology in Alzheimer’s disease and tauopathies. Acta Neuropathol. 128, 679–689 (2014).
Wang, W. et al. Serine phosphorylation of IRS1 correlates with Aβ-unrelated memory deficits and elevation in Aβ level prior to the onset of memory decline in AD. Nutrients https://doi.org/10.3390/nu11081942 (2019).
Prasetyanti, P. R. & Medema, J. P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 16, 1476–4598 (2017).
Rios, A. C. et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35, 618–632 (2019).
Hillman, Y. et al. MicroRNAs affect complement regulator expression and mitochondrial activity to modulate cell resistance to complement-dependent cytotoxicity. Cancer Immunol. Res. https://doi.org/10.1158/2326-6066.CIR-18-0818 (2019).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Ben-Hamo, R. et al. Predicting and affecting response to cancer therapy based on pathway-level biomarkers. Nat. Commun. https://doi.org/10.1038/s41467-020-17090-y (2020).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
We thank D. Douek, I. Ben-Porath and O. Sandler for their comments. We thank Champions Oncology for providing HNSCC patient tumor microarrays and PDX data. We thank M. Berger for assistance with the preparation of the figures. R.S. was partially funded by the Fabrikant-Morse Families Research Fund for Humanity, the Moross Integrated Cancer Center and the Rising Tide Foundation. R.S. is the incumbent of the Roel C. Buck Career Development Chair. I.H. was partially funded by Leona M. and Harry B. Helmsley Charitable Trust grant no. 2012PG-ISL013. I.H. and S.M.S. were partially funded by the Rabin Medical Center–Bar-Ilan grant project. G.F. is the incumbent of the David and Stacey Cynamon Research fellow Chair in Genetics and Personalized Medicine.
Author information
Authors and Affiliations
Contributions
Conceptualization was provided by A.J.B. and R.S. Experimental design and performance was conducted by A.J.B., E.G., Z.M., N.G., Y.Z., A.P., S.G., H.R., L.K., G.B., E.M., S.M.S., M.M., Y.R., J.B., I.K. and R.S. Data analysis was performed by A.J.B., E.G., Z.M., I.H., Y.R., G.F., T.G., Y.L., A.T., H.R. and R.S. Modeling was conducted by S.K.S. and Y.P. Mathematical analysis was performed by U.H. and B.B. A.J.B. and R.S. were responsible for writing the original draft of the manuscript. Supervision was carried out by R.S.
Corresponding author
Ethics declarations
Competing interests
R.S. and N.G. serve as external consultants to CuResponse, a company that utilizes ex vivo organ culture technology and have been issued equity incentives in the company. H.R. and L.K. are employees of TyrNovo, a company that develops NT219 as a potential anticancer agent, are co-inventors on NT219 patent/patent applications and have been issued equity incentives in the company. I.H. and S.M.S. are consultants of TyrNovo and have been issued equity incentives in the company.
Additional information
Peer review information Nature Cancer thanks Yoosik Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The relationship between IC50 and frequency of persisters.
The graph presents hypothetical data to demonstrate one of the differences between IC50 and frequency of persisters: the different reference point that is used to calculate these measures. IC50 is calculated by comparing the number of cells that remain viable after treatment to the number of cells in the non-treated control. The IC50 thus represents the effect of the drug on both growth rate and cell death. As opposed to IC50, the frequency of persisters is measured by comparing the number of cells that remain viable after treatment to the number of cells that were present just before drug treatment was started. By doing so, we ignore the growth inhibition effect and focus on the cell death effect.
Extended Data Fig. 2 DRPs have higher percentages of persisters than drug-naive population.
(a) The ratio of percent of persisters between PC9 DRPs that were generated by 9 days of gefitinib treatment and then been on drug holiday for 4–10 weeks and PC9 naïve cells is presented for two cell lines treated with the indicated drugs. Data are presented as mean values, bars represent the standard deviation of n = 4 independent cultures for PC9 or n = 3 independent cultures for G361. P-values were obtained by two tailed Student’s t-test. (b) The CTPgefitinib of the naïve PC9 population and selected 10 of its sub-clones as measured after 9 days of treatment. CTP of the sub-clones was normalized to the CTP of the naïve PC9 non-clonal population. Data are presented as mean values, error bars represent the standard deviation of n = 3 independent cultures. P-values were obtained by two tailed Student’s t-test. (c) PC9 clones were seeded on day 0 at 1*10^6 cells per 10 cm plate. Gefitinib or DMSO were added on day 1 at 500 nM. Floating and adherent cells were collected 48 h later, fixed and stained with anti cleaved-CASP3 (SCT #9664) followed by anti-rabbit alexa647 secondary antibody, according to the antibody manufacturer commended protocol. Flow cytometry data was acquired and analyzed by CytoFLEX LX. P-value was calculated by two tailed Student’s t-test. (d) The CTPosimertinib of PC9 clones with known high or low CTPgefitinib was measured after 7 days of treatment. CTP was normalized to the number of cells just before the initiation of treatment. Data are presented as mean values, error bars represent the standard deviation of n = 2 independent cultures. (e,f) Histograms of the CTP distribution of PC9 (e) and G361 (f) cell lines are presented for both the drug-naïve populations (blue) as well as for DRPs of these cell lines (red). CTP of clones was measured after 7 days of treatment with gefitinib. To generate DRP clones, single cells that survived 9 days of gefitinib treatment were let to proliferate into clones without drug. A right shift in the distribution is demonstrated for the DRPs. Modeling of the expected distribution by pure selection of clones according to their CTP is demonstrated in grey.
Extended Data Fig. 3 CTP does not correlate with clonal population doubling time.
Scatter plot demonstrating the CTP and clonal population doubling time of 281 naive PC9 clones (a) n = 164 PC9 DRPs (b) n = 80 naive G361 clones (c), and n = 65 G361 DRPs (d). Single cells from both cell lines were plated on 1536-well plates and left to grow for 11–16 days. To calculate the clonal population doubling time, we counted the number of cells in each well by microscopy at different time points. CTP was measured after 7 days of treatment. Due to short measurement time (24 hours), we excluded clones with PDT > 100 hours from the analysis. (e,f) Scatter plot demonstrating the CTP after 7 days of treatment and clone size before treatment for PC9 clones (n = 544 clones) (e) and G361 clones (n = 459 clones) (f). (g) Gating scheme demonstrating the measurements taken to obtain the cell-cycle distribution presented in Fig. 3f.
Extended Data Fig. 4 A wide range of CTPs exist with respect to many drugs.
(a) Single-cell derived clones of PC9 (n = 43 clones) as well as the non-clonal population were treated with the indicated drugs and concentrations for 7 days. CTP of each clone was normalized to the CTP of the non-clonal population. For each of the drugs, the clones are ordered based on their CTP (b) CTP of HCC2935 DRPs normalized to HCC2935 drug-naïve cells was measured with respect to 7 anti-cancer drugs after 7 days of treatment. HCC2935 DRPs were generated by 0.5uM gefitinib treatment for 11 days followed by drug holiday for 2–3 weeks until cells regained their normal proliferation rate. Error bars represent the standard deviation of n = 3 independent cultures.
Extended Data Fig. 5 CTP does not correlate with clonal karyotype changes or specific clonal morphology.
(a) Clonal CTP does not correlate with the total number of chromosomes. The average number of chromosomes per PC9 clone was calculated based on karyotype profiling of 10 cells/clone. Chromosomes with translocations were counted as belonging to the main chromosome that constitutes the new chromosomal derivative. Data are presented as mean values, error bars represent standard deviation of n = 10 independent cultures (b) Clonal CTP does not correlate to the individual number of chromosomes in each clone. The average number of chromosomes per PC9 clone was calculated based on karyotype profiling of 10 cells/clone. (c) Clonal CTP does not correlate with specific chromosomal derivatives (der). The average number of chromosomal aberrations per clone was calculated based on karyotype profiling of 10 cells/clone. (d) Representative images of selected PC9 clones with a wide range of CTPGefitinib (measured after 7 days of treatment) that underwent expansion to generate cell lines are presented. (e) Representative images of drug-naïve cells and DRPs of HCC2935 and H3122 cell lines. HCC2935 DRPs were generated by 0.5uM gefitinib treatment for 11 days and H3122 DRPs were generated by 0.5uM crizotinib treatment for 9 days followed by drug holiday for 2–3 weeks until cells regained their normal proliferation rate.
Extended Data Fig. 6 CTP does not correlate with pIGF1R, pEGFR or the expression of other proteins that were reported to be enriched in persisters.
(a) A scatter plot demonstrating the correlation between CTPgefitinib after 7 treatment days and averaged expression of genes that were previously suggested to be enriched in persisters. Correlation was tested across seven PC9 clones as well as the non clonal population. Each clone was sequenced in n = 3 independent cultures. (b) Levels of phosphor-EGFR were analyzed by western blot in high and low CTP clones (representative blot of n = 2 independent cultures is demonstrated). (c) Non-clonal PC9 population was subjected to combination treatment of four IGF1R inhibitors or DMSO control, with or without 1 μM gefitinib. Survival of each combination was normalized to the survival of single treatment with DMSO or gefitinib. (d) Protein levels of phosphor-IGF1-R were assayed by western blot analysis pre (upper panel) and 6 hours after (lower panel) treatment with 0.5 µM gefitinib (representative blot of n = 3 independent cultures is demonstrated). (e) Protein levels of phospho-EGFR and phospho-IGF1R were assayed by western blot analysis in PC9 drug-naïve cells or PC9 DRPs treated with 0.5uM gefitinib or DMSO control for 24 hours (representative blot of n = 2 independent cultures is demonstrated). To prepare the DRPs, PC9 drug-naïve cells were treated for 4 to 15 days with 0.5uM gefitinib, and then released from drug for additional 2–3 weeks until cells regained their normal proliferation rate.
Extended Data Fig. 7 Transcriptome and CTP analysis in RNAseq of PC9 single cell derived clones.
(a) Distributions of total molecules (UMIs) per clone without drug treatment (DMSO) and with gefitinib drug treatment (+drug). Box plots show medians as central lines. Edges of each box represent the interquartile range (IQR) and whiskers mark either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 * IQR). No removal of outliers was performed in this plot. (b) log2 of total UMIs obtained per clone in both DMSO (x axis) and drug treatment (y axis). n = 195 single-cell derived clonal populations were sampled by at least 10 K UMIs in both treatments. (c) Distribution of clonal CTP values, as measured for n = 195 clones covered in both treatments. (d) gene-gene correlation matrix, showing anti-correlation of two proliferation associated gene modules both in DMSO treated clones (upper triangle) and drug treated clones (lower triangle). (e) Total normalized expression per clone (UMIs per 100 K UMIs) summed from 200 top correlated genes to RPL8 (x axis) and RPL23 (y axis). Color of dots indicates two culturing batches of clones. (f) Expression score per clones for gene modules described in D, computed as log2 ratio of RPL8 / RPL23 total expression (x axis), and is not associated with clonal CTP (y axis). Normalized CTP per clone was computed as log2 ratio of each clone’s CTP to median CTP of the relevant culturing batch. (g) Expression of RPL8 gene module is positively correlated with growth rates. Clones are binned according to the percentage of increment in cells, measured between 5 and 7 days since splitting cells to growth assay (see Methods), and RPL8 module expression is computed as summation of clonal UMIs for 10 genes with highest correlation to RPL8 (units of UMIs per 100 K UMIs). (h) Scatter plots showing the normalized expression of Mucin5B pre- and 24 hours post- 0.5 μM gefitinib treatment. Spearman’s correlation coefficient is indicated for each plot. (i) Cumulative distributions of Spearman’s correlation of individual gene expression to CTP in DMSO treated clones. (Top-left) Showing lowest correlation values obtained between genes and CTP (red), and lowest correlation between genes and a randomized vector of CTP values (orange). (Top-right) Showing highest correlation between genes and CTP (blue), and between genes and a randomized vector of CTP values (lightblue). (Bottom panel) Displaying genes with highest (blue) and lowest (red) correlation to CTP, along with their correlation to randomized CTP vector (lightblue and orange bars). (j) as h, for gefitinib treated clones. Highlighting genes that maintain strong correlation to CTP in both drug treated and untreated clones. (k) Scatter plots showing log normalized expression of selected genes (x axis) and clonal CTP (y axis). (l) Scatter plots showing the normalized expression of ALDH1A3 pre- and 24 hours post- 0.5 μM gefitinib treatment. Spearman’s correlation coefficient is indicated for each plot. Statistical significance for H and L was assessed using a two-tailed Student’s t-test.
Extended Data Fig. 8 Gene modules and CTP analysis in RNAseq of PC9 single-cell derived clones.
(a) gene-gene correlation matrix, showing gene modules unrelated to proliferation gradient defined in Fig.S7d-g, computed based on normalized expression in untreated clones (upper triangle) and in gefitinib treated clones (lower triangle). (b) log normalized clonal expression of gene modules specified in A in untreated (x axis) and drug treated clones (y axis). Dashed light blue line defining the diagonal, indicating changes of absolute expression levels in untreated and treated clones. A higher correlation suggests higher clonal stability of the gene module, amid the transcriptional response to the drug treatment. (c) Showing 50 genes with highest (blue) and lowest (red) fold-change in Mucin-high clones vs Mucin-low clones. Fold-change computed as log ratio of expression geometric mean of each gene in 30% of gefitinib treated clones with highest Mucin expression, to its expression geometric mean in 30% of gefitinib treated clones with lowest Mucin expression. (d) Clonal expression of Mucin gene module is associated with CTP. Showing expression of genes associated with the Mucin and ZNF469 genes modules shown in A for gefitinib treated (upper panel) and DMSO treated clones (lower panel). Each column in both panel defines clone from the same single-cell-derived clonal population, after eliminating one of the n = 195 clones whose CTP-assay failed technically. Expression of each gene is normalized to maximal expression in gefitinib treated clones. Despite the weaker expression in untreated clones, the conserved expression structure which correlated to CTP can be observed also without treatment. (e) Average of the mucin gene module score is presented for clones that were grouped by their CTP gefitinib. P-values were calculated using the chi-square test.
Extended Data Fig. 9 IRS1 phosphorylation and response to therapy.
(a) Pearson’s correlation determination (r^2) test was used to calculate the correlation coefficient and the p-value between CTP and all detected phosphorylation sites in IRS1 (upper panel) and IRS2 (lower panel). The correlation was calculated based on n = 7 clones as well as the non-clonal PC9 population. FDR < 0.1 (b) pIRS1 (ser1101) and pIRS1 (ser1078) were measured in drug-naïve cells and DRPs of the G361 cell line treated 0.06 µM trametinib. Data are presented as mean values, error bars represent the standard deviation of n = 3 independent cultures. p-values were calculated by one tailed Student’s t-test.
Extended Data Fig. 10 CTP and the long term impact of competing treatments.
While the mean CTP captures a treatment’s short term impact, the complete CTP distribution, determines its long term effect. (a) Two competing hypothetical drugs (blue) and (red) are demonstrated. While the mean CTP of drug B (distribution peak) is smaller, indicating a rapid decline in the cancer cell population within the first week, the tail of this distribution is broader, and hence it exhibits a higher variance in CTP compared to drug A. (b) The calculated cell count, vs. time under the two treatments is presented. At day 7, drug B seems superior, however, as a result its broad tailed distribution, after approximately 2 weeks of treatment it becomes less effective than drug A. Such crossover illustrates the importance of the CTP distribution, as opposed to just the mean, as a predictor of treatment effectiveness. (c) An empirical example showing the cancer cell population under doxorubicin (blue) vs. trametinib (red). While the latter is three times more effective at day 7, the trend is reversed in later time points. (d) Similar crossover is observed for cisplatin vs. trametinib.
Supplementary information
Supplementary Video 1
Time-lapse video following PC9 cells treated with 0.5 µM gefitinib for 80 d.
Supplementary Video 2
Time-lapse video following PC9 cells treated with 0.5 µM gefitinib for 80 d.
Supplementary Video 3
Time-lapse video following PC9 cells treated with 0.5 µM gefitinib for 80 d.
Supplementary Tables
Supplementary Tables 1–8.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Jacob Berger, A., Gigi, E., Kupershmidt, L. et al. IRS1 phosphorylation underlies the non-stochastic probability of cancer cells to persist during EGFR inhibition therapy. Nat Cancer 2, 1055–1070 (2021). https://doi.org/10.1038/s43018-021-00261-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00261-1
This article is cited by
-
Methylation across the central dogma in health and diseases: new therapeutic strategies
Signal Transduction and Targeted Therapy (2023)
-
Drug-tolerant persister cells in cancer: the cutting edges and future directions
Nature Reviews Clinical Oncology (2023)
-
A heritable, non-genetic road to cancer evolution
Nature (2022)