Abstract
Chemotherapy is a standard treatment for pediatric acute lymphoblastic leukemia (ALL), which sometimes relapses with chemoresistant features. However, whether acquired drug-resistance mutations in relapsed ALL pre-exist or are induced by treatment remains unknown. Here we provide direct evidence of a specific mechanism by which chemotherapy induces drug-resistance-associated mutations leading to relapse. Using genomic and functional analysis of relapsed ALL we show that thiopurine treatment in mismatch repair (MMR)-deficient leukemias induces hotspot TP53 R248Q mutations through a specific mutational signature (thio-dMMR). Clonal evolution analysis reveals sequential MMR inactivation followed by TP53 mutation in some patients with ALL. Acquired TP53 R248Q mutations are associated with on-treatment relapse, poor treatment response and resistance to multiple chemotherapeutic agents, which could be reversed by pharmacological p53 reactivation. Our findings indicate that TP53 R248Q in relapsed ALL originates through synergistic mutagenesis from thiopurine treatment and MMR deficiency and suggest strategies to prevent or treat TP53-mutant relapse.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
There was no active patient recruitment or prospective clinical trial data for the present study, but rather this was a reanalysis of existing datasets and clinical and genomics data were obtained as follows. Trials registered at ClinicalTrials.gov were AALL0232 (ref. 56) (NCT00075725), AALL0331 (ref. 57) (NCT00103285), ALL-BFM-2000 (ref. 59) (NCT00430118), COALL 07-03 (ref. 61) (NCT00343369) and ALL-REZ BFM 2002 (ref. 22) (NCT00114348). A trial registered at the Chinese Clinical Trial Registry was CCCG-ALL-2015 (ref. 55) (ChiCTR-IPR-14005706). Trials not registered at ClinicalTrials.gov or at an equivalent agency (initiated before ClinicalTrials.gov release in 2000) were XH-ALL-99 (ref. 53) (China), SCMC-ALL-2005/2009 (refs. 51,52) (China), CCLG-ALL-2008 (ref. 54) (China), ALL-BFM 95 (ref. 62) (Germany), COALL 06-97 (ref. 60) (Germany) and ALL-REZ BFM 95/96 (ref. 58) (Germany). TARGET is a cooperative collaboration, initiated in 2006, between NCI, the Children’s Oncology Group, University of Colorado Cancer Center, University of New Mexico Cancer Center and St. Jude Children’s Research Hospital (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000218.v23.p8). TARGET raw sequencing data are available through dbGaP using accession phs000218. Data from ‘TARGET’ datasets need controlled access to ensure patient data protection and requests to access these datasets should be directed to ‘dbGaP web portal’ where access will be supervised by the NCI Data Access Committee. PCGP data raw data are available at www.stjude.cloud under controlled access to protect patient data; data access requests are reviewed by the PCGP Steering Committee. PCGP raw data were originally deposited on European Genome–phenome Archive (EGAD00001000261, EGAD00001001864, EGAD00001000260, EGAD00001000161, EGAD00001000259, EGAD00001000164, EGAD00001000160, EGAD00001000163, EGAD00001000268, EGAD00001000162, EGAD00001000085, EGAD00001000165, EGAD00001000135, EGAD00001000159) and reanalyzed using .bam files and somatic VCF files obtained from St. Jude Cloud Genomics Platform (https://platform.stjude.cloud/data/cohorts#) by selecting the study ‘Pediatric Cancer Genome Project (PCGP).’ TCGA/PCAWG SNV and indel data were obtained through the International Cancer Genome Consortium Data Portal at https://dcc.icgc.org/ by using the ‘Advanced Search’ option, selecting ‘PCAWG’ for ‘Study’, ‘WGS’ for ‘Donor Analysis Type’ and ‘SSM’ (SNVs and indels) for ‘Available Data Type’. These data are available without controlled access and no accession number is needed. TP53 mutation data in different cancer types were downloaded from cBioPortal. ChIP-seq data were submitted to the Genome Sequence Archive with accession code HRA000386. All other data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
Custom code used in this study for routine data processing and formatting is available at https://github.com/samuelwb1985/TP53. No algorithms or code packages were developed in this study.
References
Bouaoun, L. et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum. Mutat. 37, 865–876 (2016).
Levine, A. J. & Oren, M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758 (2009).
Zhou, B. B. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000).
Li, B. et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 135, 41–55 (2020).
Hof, J. et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 29, 3185–3193 (2011).
Wong, T. N. et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 518, 552–555 (2015).
Diouf, B. et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat. Med. 17, 1298–1303 (2011).
Waanders, E. et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov. 1, 96–111 (2020).
Ma, X. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018).
Downing, J. R. et al. The Pediatric Cancer Genome Project. Nat. Genet. 44, 619–622 (2012).
Andersson, A. K. et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 47, 330–337 (2015).
Liu, Y. et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 49, 1211–1218 (2017).
Zhang, J. et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 48, 1481–1489 (2016).
Roberts, K. G. et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 371, 1005–1015 (2014).
Holmfeldt, L. et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45, 242–252 (2013).
Eckert, C. et al. Improving stratification for children with late bone marrow B-cell acute lymphoblastic leukemia relapses with refined response classification and integration of genetics. J. Clin. Oncol. https://doi.org/10.1200/JCO.19.01694 (2019).
Groeneveld-Krentz, S. et al. Aneuploidy in children with relapsed B-cell precursor acute lymphoblastic leukaemia: clinical importance of detecting a hypodiploid origin of relapse. Br. J. Haematol. 185, 266–283 (2019).
Hof, J. et al. NOTCH1 mutation, TP53 alteration and myeloid antigen expression predict outcome heterogeneity in children with first relapse of T-cell acute lymphoblastic leukemia. Haematologica 102, e249–e252 (2017).
Oshima, K. et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 113, 11306–11311 (2016).
Stengel, A. et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 124, 251–258 (2014).
Wada, M. et al. Analysis of p53 mutations in a large series of lymphoid hematologic malignancies of childhood. Blood 82, 3163–3169 (1993).
Eckert, C. et al. Use of allogeneic hematopoietic stem-cell transplantation based on minimal residual disease response improves outcomes for children with relapsed acute lymphoblastic leukemia in the intermediate-risk group. J. Clin. Oncol. 31, 2736–2742 (2013).
Yu, C. H. et al. TP53 alterations in relapsed childhood acute lymphoblastic leukemia. Cancer Sci. 111, 229–238 (2020).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Alexandrov, L. B. et al. Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015).
Evensen, N. A. et al. MSH6 haploinsufficiency at relapse contributes to the development of thiopurine resistance in pediatric B-lymphoblastic leukemia. Haematologica 103, 830–839 (2018).
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Holt, S. M., Scemama, J. L., Panayiotidis, M. I. & Georgakilas, A. G. Compromised repair of clustered DNA damage in the human acute lymphoblastic leukemia MSH2-deficient NALM-6 cells. Mutat. Res. 674, 123–130 (2009).
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J. & Stratton, M. R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013).
Boot, A. et al. In-depth characterization of the cisplatin mutational signature in human cell lines and in esophageal and liver tumors. Genome Res. 28, 654–665 (2018).
You, C., Dai, X., Yuan, B. & Wang, Y. Effects of 6-thioguanine and S6-methylthioguanine on transcription in vitro and in human cells. J. Biol. Chem. 287, 40915–40923 (2012).
Morganella, S. et al. The topography of mutational processes in breast cancer genomes. Nat. Commun. 7, 11383 (2016).
Bykov, V. J. N. et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288 (2002).
Lambert, J. M. R. et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388 (2009).
Demir, S. et al. Therapeutic targeting of mutant p53 in pediatric acute lymphoblastic leukemia. Haematologica 105, 170–181 (2020).
Lehmann, S. et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J. Clin. Oncol. 30, 3633–3639 (2012).
Zarin, D. A., Tse, T., Williams, R. J. & Rajakannan, T. Update on trial registration 11 years after the ICMJE policy was established. New Engl. J. Med. 376, 383–391 (2017).
Chen, Y. L. et al. Blocking ATM-dependent NF-κB pathway overcomes niche protection and improves chemotherapy response in acute lymphoblastic leukemia. Leukemia 33, 2365–2378 (2019).
Duan, C. W. et al. Leukemia propagating cells rebuild an evolving niche in response to therapy. Cancer Cell 25, 778–793 (2014).
Swann, P. F. et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science 273, 1109–1111 (1996).
Uribe-Luna, S. et al. Mutagenic consequences of the incorporation of 6-thioguanine into DNA. Biochem. Pharmacol. 54, 419–424 (1997).
Schwartz, J. R. et al. The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms. Nat. Commun. 12, 985 (2021).
Wang, H. & Wang, Y. 6-Thioguanine perturbs cytosine methylation at the CpG dinucleotide site by DNA methyltransferases in vitro and acts as a DNA demethylating agent in vivo. Biochemistry 48, 2290–2299 (2009).
Lans, H., Hoeijmakers, J. H. J., Vermeulen, W. & Marteijn, J. A. The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 20, 766–784 (2019).
Hanawalt, P. C. & Spivak, G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008).
Teng, Y. et al. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat. Commun. 9, 4115 (2018).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Steuer, C. E. & Ramalingam, S. S. Tumor mutation burden: leading immunotherapy to the era of precision medicine? J. Clin. Oncol. 36, 631–632 (2018).
Zamora, A. E. et al. Pediatric patients with acute lymphoblastic leukemia generate abundant and functional neoantigen-specific CD8(+) T cell responses. Sci. Transl. Med. 11, eaat8549 (2019).
Chang, T. C. et al. The neoepitope landscape in pediatric cancers. Genome Med. 9, 78 (2017).
Liang, Y. et al. Treatment outcome of young children with acute lymphoblastic leukaemia: achievements and directions implied from Shanghai Children’s Medical Centre based SCMC-ALL-2005 protocol. Br. J. Haematol. 169, 267–277 (2015).
Liu, K. et al. Long-term follow-up of acute lymphoblastic leukemia in young children treated by the SCMC-ALL-2009 protocol. Leuk. Lymphoma 61, 2850–2858 (2020).
Gu, L. J. et al. Clinical outcome of children with newly diagnosed acute lymphoblastic leukemia treated in a single center in Shanghai, China. Leuk. Lymphoma 49, 488–494 (2008).
Hu, Y. X. et al. A prospective evaluation of minimal residual disease as risk stratification for CCLG-ALL-2008 treatment protocol in pediatric B precursor acute lymphoblastic leukemia. Eur. Rev. Med. Pharmacol. Sci. 20, 1680–1690 (2016).
Cai, J. et al. Treatment abandonment in childhood acute lymphoblastic leukaemia in China: a retrospective cohort study of the Chinese Children’s Cancer Group. Arch. Dis. Child. 104, 522–529 (2019).
Larsen, E. C. et al. Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: a report from Children’s Oncology Group study AALL0232. J. Clin. Oncol. 34, 2380–2388 (2016).
Maloney, K.W. et al. Outcome in children with standard-risk B-cell acute lymphoblastic leukemia: results of Children’s Oncology Group trial AALL0331. J. Clin. Oncol. https://doi.org/10.1200/JCO.19.01086 (2020).
Eckert, C. et al. Minimal residual disease after induction is the strongest predictor of prognosis in intermediate risk relapsed acute lymphoblastic leukaemia: long-term results of trial ALL-REZ BFM P95/96. Eur. J. Cancer 49, 1346–1355 (2013).
Conter, V. et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 115, 3206–3214 (2010).
Escherich, G. et al. The long-term impact of in vitro drug sensitivity on risk stratification and treatment outcome in acute lymphoblastic leukemia of childhood (CoALL 06-97). Haematologica 96, 854–862 (2011).
Schramm, F. et al. Results of CoALL 07-03 study childhood ALL based on combined risk assessment by in vivo and in vitro pharmacosensitivity. Blood Adv. 3, 3688–3699 (2019).
Moricke, A. et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood 111, 4477–4489 (2008).
Tate, J. G. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019).
Armbruster, D. A. & Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29, S49–S52 (2008).
Edmonson, M. N. et al. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics 27, 865–866 (2011).
Chen, X. et al. CONSERTING: integrating copy-number analysis with structural-variation detection. Nat. Methods 12, 527–530 (2015).
Wang, J. et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods 8, 652–654 (2011).
Li, B. et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat. Med. 21, 563–571 (2015).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Miller, C. A. et al. Visualizing tumor evolution with the fishplot package for R. BMC Genomics 17, 880 (2016).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31530017, 81861148026 and 81970141 to B.-B.S.Z.; 81800145 to F.Y.; 81870116 and 82070159 to H.L.; 82000110 to C.T.; and 81670174 to B.L.), Shanghai Key Laboratory of Clinical Molecular Diagnostics for Pediatrics (20dz2260900 to B.-B.S.Z.), the National Key R&D Program of China, Stem Cell and Translation Research (no. 2016YFA0102000 to C.-W.D.), Rising-Star Program from the Science and Technology Commission of Shanghai Municipality (18QA1402900 to H.L.), Shanghai Committee of Science and Technology (21ZR1441000 to F.Y.), the German Childhood Cancer Foundation (DKS-2015.07 to R.K.-S. and C.E.; DKS-2007.02 to A.v.S.) and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. The authors acknowledge the International Cancer Genome Consortium and TCGA for access to PCAWG data and the National Institutes of Health/NCI for use of the TARGET datasets. We thank Shanghai Lide Biotech Co. Ltd. for laboratory animal services, Genechem Co. Ltd. for plasmid construction and virus packaging services, Shanghai Gemple Biotech Co. Ltd for the Biorad QX200 ddPCR equipment, Qiaochu Chen for generating R158H and R158C constructs and performing some of the related western blot and cell viability experiments, J. Huang and Y. An (Shanghai Jiao Tong University) for Raindance ddPCR equipment and experimental advice, S. Liu (Fudan University) for MCF10A culture medium, P. Qian (Zhejiang University School of Medicine) for the bioluminescence plasimd. We are grateful to G. Zambetti (St. Jude Children’s Research Hospital), M. Lu (Shanghai Ruijin hospital), G.-M. Li (UT Southwestern) and H. Jiang (Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences) for critical comments and constructive suggestions on the manuscript.
Author information
Authors and Affiliations
Contributions
B.-B.S.Z., R.K.-S., S.W.B., F.Y. and J.Z. designed the research strategy. S.W.B., B.-B.S.Z., R.K.-S., J.Z., F.Y., C.T., H.S. and Y.L. wrote the manuscript and organized the figures. B.L., S.S., J.C., L. Ding, H.S. T.W. and H.L. collected Chinese cohort samples and analyzed Chinese cohort clinical data. M.J.B., R.K.-S., J.H. A.v.S. and C.E. collected German cohort samples and analyzed German cohort clinical data. S.W.B., X.M., Y.L. and J.Z. analyzed US cohort clinical and sequencing data. S.W.B., H.S., X.M., Y.L., P.K. and O.P. performed bioinformatics analysis. S.W.B. performed mutation signature analysis. J.M.K. helped thio-dMMR signature analysis in AML. Y.C. generated cells for signature analysis. F.Y. performed ddPCR assays. F.Y., H.F., X.M., H.S., B.L. and L. Ding, performed deep sequencing of minimal residual disease samples and clonal evolution analysis. F.Y., L. Du and M.J.B. generated cell lines for functional assays. F.Y., H.F., L. Du, X.L. and C.T. performed drug sensitivity assays. L. Du and F.Y. performed cell cycle and apoptosis analysis. L. Du, C.T. and X.L. performed western blot analysis. F.Y. and H.S. performed ChIP-seq analysis. C.T., H.F., C.D., H.L. and H.C. designed and performed animal xenograft and PDX experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks William Carroll, Pieter Van Vlierberghe and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Summary of seven ALL cohorts and TP53 mutations information in this study, and distribution of different TP53 R248 mutations in ALL and other cancers.
a, Diagnosis only patients are shown in dark green, diagnosis and relapse paired patients are shown in light green and orange pairs, and relapse only patients are shown in orange. See online methods for cohort details. Cohorts within dotted border had WGS data and were used for mutagenesis analysis. Stengel et al. cohort is mixed with adult and pediatric ALL, while the other six cohorts are all pediatric ALL. The numbers of patients and TP53 mutations in each cohort or each portion within a cohort are shown individually. dx stands for diagnosis, rel for relapse, mt for mutation, and pts for patients. Two indels in the Yu et al. cohort and three indels in the Stengel et al. cohort were without genome positions and therefore could not be included in further analysis. b, Age distribution in four primarily pediatric cohorts with detailed age information. Age was available for all patients from the SCMC and the ALL-REZ BFM cohorts, and for 271/277 patients from the PCGP cohort and for 218/221 patients from the TARGET cohort. c, Bar plot shows the percentage of TP53 R248Q, R248W and other types of R248 mutations (including R248P, R248G and R248L) in diagnosis ALL, relapsed ALL and other tumor types (including TCGA tumors and IARC liver cancer). d, Pie charts show percentage of TP53 R248Q, R248W and other types of R248 mutations in diagnosis ALL, relapsed ALL and other tumor types. e, Bar plots show frequency of different R248 mutations among all TP53 mutations in different cohorts. Relapsed ALL represents data from all (China, German, USA, Taiwan) cohorts combined, including both relapse-specific and relapse-shared R248 mutations.
Extended Data Fig. 2 Relapse-specificity of 28 TP53 mutations found in paired Chinese and US cohorts, and serial sample analysis revealing dynamics of TP53 mutant clone evolution in different models.
a, Pie charts show the distributions of TP53 mutations and R248Q in three clonal evolution models. b, Schemes show the allele frequency, sequencing depth and clonal evolution pattern of TP53 alterations in paired patients from Chinese and US cohorts. Most of TP53 R248Q mutation in these cohorts are relapse-specific and could be de novo acquired during chemotherapy treatment. 6 out of 8 relapse-specific R248Q mutations were confirmed by ddPCR. The TP53 R248Q in ALL105, the TP53 R267Q in ALL063, and the TP53 R175H in SJETV010 were detected at second relapse (R2) but not first relapse (R1). c, Germline model: ALL237, germline TP53 mutation with ancestral clonal evolution pattern. Graphic representation (‘fish plots’) of clonal evolution shaped by chemotherapy based on ultra-deep sequencing. Treatment history is shown at the tops with used drugs indicated by different colors. Clonal evolution is shown in the middle with the vertical axis representing proportion of cells, normal cells are represented by white space. The minimum tumor purity values (thin line) indicate no detectable ALL cells by ultra-deep sequencing, and their clonal compositions at these times inferred. Days from diagnosis are indicated at the bottom, with tick marks indicating time points at which samples were sequenced. d, Pre-existing model: ALL064, pre-existing TP53 mutation with selective clonal evolution pattern. See (c) legend for fish plot details. e, Acquired model: ALL161, acquired TP53 mutation with de novo clonal evolution pattern. See (c) legend for fish plot details.
Extended Data Fig. 3 Comparison of mutational signature spectra and mutation burden.
a, Top, average mutational spectrum of 591 diagnosis ALL WGS samples without an MMR gene mutation (error bars, SEM). Second, COSMIC signature 1. Third, the thio-dMMR signature, as defined by Nalm6 (MSH2-null) treated with 6-TG (mean of 3 6-TG treated clones’ spectra minus 3 untreated clones). Fourth, MCF10A with engineered MSH2−/− treated with 6-TG (mean of two treated clones’ spectra minus two untreated clones). ALL diagnosis samples bear COSMIC signature 1, as their C > T spectrum at NCG resembles COSMIC signature 1 (cosine similarity 0.99 of C > T at NCG). COSMIC signature 1 is similar to the thio-dMMR signature (cosine similarity 0.92 comparing their entire profiles). However, they differ in mutation rate (Fig. 2e) and strand bias, with COSMIC signature 1 having strand bias only at ACG, and the thio-dMMR signature having strand bias at all NCG positions, among C > T mutations (see asterisks and strand bias plots at right, constructed as in Fig. 2g; T indicates transcribed and U the untranscribed strand). Strand bias plots show SNVs in the top 25% of expressed genes’ bodies; y-axis represents the proportion of all SNVs within gene bodies of highly expressed genes. The diagnosis ALL plot (top-right) represents 76 diagnosis ALL samples with both WGS and RNA-Seq; only the ACG mutations had statistically significant strand bias for diagnosis ALL (P < 0.05), while all NCG had statistically significant strand bias in thio-dMMR signature-positive relapsed ALL (bottom-right). P values, two-sided paired t-test of patient samples shown (excluding Nalm6 and MCF10A). b, Boxplot showing relapse-specific SNV burden, assessed by WGS of 181 diagnosis-relapse ALL pairs, in samples lacking a relapse-specific MMR gene alteration (WT, n = 151 patients) and those bearing one (mutated, n = 30 patients). Box, interquartile range; middle bar, median. Whiskers use a 1.5*interquartile range rule, each point represents one sample. P value, two-sided Wilcoxon test.
Extended Data Fig. 4 Example mutation spectra illustrating thio-dMMR and MMR deficiency mutational signatures.
The spectra of signatures (in boxes) and samples are indicated with the trinucleotide context indicated at the bottom, and mutation type (6 mutation types, such as C > A, C > G, etc., which occur at the center of the trinucleotide context) at top. Y-axis represents the proportion of SNVs for signatures and the number of SNVs for samples. a, The thio-dMMR signature as determined from treatment of Nalm6 cells (MSH2-null) with 6-TG for 5 weeks, with the mean of the 3 treated clones’ mutational spectra subtracted from the mean of 3 untreated clones’ spectra (negative values were set to zero; top) and several example patients’ relapse-specific mutation catalogs beneath. Relapse-specific MMR mutations and, if applicable, TP53 mutations present in each sample are also indicated. b, Three COSMIC mutational signatures associated with MMR deficiency (top) and several diagnosis samples resembling one or more of these signatures plus a mutation in an MMR gene (bottom). This shows that the standard MMR signatures are distinct from the thio-dMMR signature, which has similarities to the standard thiopurine signature.
Extended Data Fig. 5 Characterization of MMR-deficient MCF10A MSH2−/− and Nalm6 cells.
a, Left, western blot showing the lack of MSH2 expression in Nalm6; right, knockout of MSH2 gene by CRISPR/Cas9 in MCF10A cells. b, Proliferation assay showing the effect of MSH2 knockout on 6-TG sensitivity of MCF10A cells. MMR-deficient Nalm6 was used for comparison. Data mean ± SD is shown, n = 3 biologically independent experiments. c, Top, mean of three untreated Nalm6 single-cell clones’ spectra after 5 weeks of culture as they diverged from bulk pre-treatment. Second, mean of two untreated MCF10A MSH2−/− clones after 15 weeks’ untreated culture. Their spectra are a combination of MMR deficiency-associated COSMIC signatures 26 (third) and 6 (fourth), with cosine similarities of the T > C and C > T spectra for those two signatures indicated, respectively, relative to Nalm6 and MCF10A. Signatures 6 and 26 lack strong strand bias of C > T mutations at NCG, in contrast to the thio-dMMR signature (right, which reproduces Fig. 2g, showing diagnosis samples with MMR gene mutations). Only C > T at ACG showed significant (though weak) strand bias. P value, two-sided paired t-test of the patient samples shown. d, Number of relapse-specific SNVs caused by each mutational signature based on paired diagnosis-relapse WGS. This is similar to Fig. 2e, with the same sample set, except that here the number of relapse-specific SNVs is shown, rather than the number of relapse-specific SNVs per day. Samples with available relapse time were analyzed (n = 142 samples); for each signature, only samples bearing the signature are shown. Box, interquartile range; center line, median. Whiskers use a 1.5*interquartile range rule. P values are by two-tailed t-test.
Extended Data Fig. 6 Comprehensive strand bias analysis and schematic of possible mechanism of the thio-dMMR signature’s strand bias.
a, This panel reproduces strand bias data in Fig. 2g and Extended Data Fig. 3, for C > T SNVs at different NCG trinucleotide contexts. Those figures show only strand bias information in SNVs in the top 25% of expressed genes (which are reproduced here, at left). This figure also shows, for comparison, SNVs in lowly-expressed genes (bottom 75% of expression by RNA-Seq) to indicate that the strand bias is indeed likely due to transcription, since it largely disappears in lowly-expressed genes (right). Data were derived from samples with both WGS (for SNV detection) and RNA-Seq (to determine whether SNVs are in highly-expressed genes or not). Y-axis represents the proportion of all SNVs within gene bodies of highly expressed (left) or lowly expressed (right) genes in each sample; T indicates transcribed while U indicates untranscribed strand. Top represents diagnosis samples to indicate COSMIC signature 1 strand bias; middle represents relapsed ALL with MMR mutations and the thio-dMMR signature as in Fig. 2g; bottom represents diagnosis samples with MMR mutations as in Fig. 2g. P values are by two-sided paired t-test of only the patient samples in each panel (excluding MCF10A and Nalm6 data). b, When thioguanine (green) is incorporated into the DNA strand that will be transcribed (blue), RNA polymerase II recognizes the aberrant thioguanine base and initiates transcription-coupled repair to replace it with a standard guanine residue15, leading to no mutation. c, When thioguanine is incorporated into the DNA strand that will not be transcribed (gray), RNA polymerase II does not initiate transcription-coupled repair30. After subsequent DNA replication, the unrepaired thioguanine will be mispaired with thymine25. This leads to a G > A mutation on the untranscribed strand, and C > T on the transcribed strand. Because mutational signature analysis by convention considers the pyrimidine (not purine) reference allele for each point mutation, G > A mutations are considered C > T mutations. This is consistent with the strand bias in Fig. 2g, top. If, however, the strand bias in Fig. 2g were reversed, we would assume that mutagenesis initiated at cytosine rather than guanine in guanine-cytosine base pairs.
Extended Data Fig. 7 Example calculation of the probability that a TP53 SNV was caused by a specific mutational signature.
Top panel shows an example TP53 variant that was likely thiopurine-induced. Left shows an example relapse-specific mutation catalog, in this case for ALL245, which harbors a relapse-specific TP53 R248Q variant (a C > T mutation at context CCG) and the thiopurine signature. Right shows how this relapse-specific catalog was deconvoluted into three signatures, which when added together are very similar (cosine similarity of 0.95) to the actual sample mutation catalog. Each signature’s dosage is indicated as the number of mutations caused by the signature in parenthetical text. The mutation spectra at right represent the absolute numbers of mutations at each mutation channel which are predicted to have been caused by each signature. The C > T (CCG) channel had some 64.9 mutations caused by the thiopurine signature, while the other signatures caused 16.2 and 5.8 mutations (signatures 1 and 5, respectively). The calculation at the bottom shows that there is therefore a 74.7% likelihood that the TP53 R248Q mutation was caused by thiopurines, as the thiopurine signature contributed that percentage of mutations occurring at the TP53 R248Q channel (C > T at CCG). Bottom shows another example where the TP53 variant was likely induced by signature 1, using a diagnosis glioblastoma (GBM) sample from the TCGA.
Extended Data Fig. 8 TP53 R248Q and MSH6 Q949P mutations are in the same sub-clone of ALL105 second relapse sample.
The sanger sequencing results of the bulk DNA and one single cell DNA of PDXs derived from ALL105 second relapse sample (R2) are shown with red arrows pointing the mutated nucleotides in TP53 and MSH6 genes respectively. The bulk TP53 and MSH6 VAFs may be increased here compared to the clonal evolution diagram in Fig. 3c due to selection for the TP53 and MSH6 clone in the PDX.
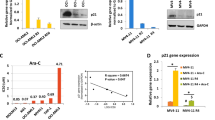
Extended Data Fig. 9 Chemotherapeutic drug sensitivities, DDR and p53 pathway biomarkers, and p53 pathway target genes analysis of p53 mutants in relapsed ALL.
a, Schematic showing the construction of TP53 knockout and re-expression of different p53 mutants in BCP-ALL cell lines. b, Thiopurine (6MP and 6-TG) drug and the non-DNA damaging compounds (dexamethasone, DEX and L-asparaginase, L-ASP) drug sensitivities (IC50s) of TP53 knockout and re-expression of p53 mutants in cells derived from Nalm6. Data are shown as mean ± SD, n = 3 biologically independent experiments. c, Snapshots of p53 ChIP-seq data of TP53 knockout and re-expression cells derived from Nalm6 at the CDKN1A, BBC3/PUMA and MDM2 gene loci. The RefSeq genes are given below the binding peaks. The genome-wide data are plotted in reads per million mapped reads (R.P.M.). d, The transcription levels of CDKN1A, BBC3/PUMA and MDM2 relative to the ACTB gene in TP53 knockout and re-expression cells derived from Nalm6 measured by real-time RT-PCR. Data are shown as mean ± SD, n = 3 biologically independent experiments. e, Nalm6 derived cells and (f) Reh derived cells after treatment with 1 μM idarubicin for 6 hours. Actin was used as a loading control.
Extended Data Fig. 10 APR-246 decreases the chemotherapy resistance of TP53 R248Q ALL cells and increases chemo-induced apoptosis in primary ALL cells with R248Q mutation.
a, The IC50s of idarubicin with (black) or without (white) treatment of 10 μM APR-246 in TP53 knockout and re-expression cells derived from Nalm6 cells. Data are shown as mean ± SD, n = 3 biologically independent experiments. b, The IC50s of idarubicin with (black) or without (white) treatment of 10 μM APR-246 in TP53 knockout and re-expression cells derived from Reh cells. Data are shown as mean ± SD, n = 3 biologically independent experiments. c, Western blots of p21 and PUMA in TP53 knockout and re-expression cells derived from Reh with or without 30 μM APR-246 pre-treatment for 12 hours followed by 1 μM idarubicin treatment for 6 hours. Actin was used as a loading control. d, Primary cultured cells derived from matched diagnosis and relapsed ALL samples with acquired TP53 mutations at relapse were treated with 1 μM idarubicin or/and 30 μM APR-246 treated for 12 hours, then apoptosis was detected by Annexin V staining. e, The statistical bar chart of (d). Data are shown as mean ± SD, n = 3 biologically independent experiments.
Supplementary information
Supplementary Information
Supplementary Fig. 1, exemplifying the gating strategy.
Supplementary Tables
Supplementary Tables 1–5
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 10
Statistical source data.
Source Data Extended Data Fig. 10
Unprocessed western blots and/or gels.
Rights and permissions
About this article
Cite this article
Yang, F., Brady, S.W., Tang, C. et al. Chemotherapy and mismatch repair deficiency cooperate to fuel TP53 mutagenesis and ALL relapse. Nat Cancer 2, 819–834 (2021). https://doi.org/10.1038/s43018-021-00230-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00230-8
This article is cited by
-
Relapsed Acute Lymphoblastic Leukemia
Indian Journal of Pediatrics (2024)
-
Targeting DNA polymerase β elicits synthetic lethality with mismatch repair deficiency in acute lymphoblastic leukemia
Leukemia (2023)
-
Clonal evolution in leukemia: preleukemia, evolutionary models, and clinical implications
Genome Instability & Disease (2023)
-
In vivo PDX CRISPR/Cas9 screens reveal mutual therapeutic targets to overcome heterogeneous acquired chemo-resistance
Leukemia (2022)
-
Detection of early seeding of Richter transformation in chronic lymphocytic leukemia
Nature Medicine (2022)