Abstract
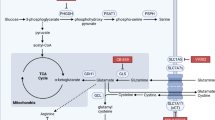
Brain metastases are refractory to therapies that control systemic disease in patients with human epidermal growth factor receptor 2-positive breast cancer and the brain microenvironment contributes to this therapy resistance. Nutrient availability can vary across tissues, therefore metabolic adaptations required for brain metastatic breast cancer growth may introduce liabilities that can be exploited for therapy. Here we assessed how metabolism differs between breast tumors in brain versus extracranial sites and found that fatty acid synthesis is elevated in breast tumors growing in the brain. We determine that this phenotype is an adaptation to decreased lipid availability in the brain relative to other tissues, resulting in site-specific dependency on fatty acid synthesis for breast tumors growing at this site. Genetic or pharmacological inhibition of fatty acid synthase reduces human epidermal growth factor receptor 2-positive breast tumor growth in the brain, demonstrating that differences in nutrient availability across metastatic sites can result in targetable metabolic dependencies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Previously published microarray and RNA-seq data that were re-analyzed here are available in GEO under accession codes GSE86849 and GSE14020 and RNA-seq data that were re-analyzed here are available at https://github.com/npriedig/jnci_2018.
Metabolomics data have been deposited to the EMBL-EBI MetaboLights database73 with the identifier MTBLS2434. The complete dataset can be accessed here https://www.ebi.ac.uk/metabolights/MTBLS2434.
The data generated for this study are included in the manuscript, extended data, supplementary files and the provided source data files. Source data that have not been provided are available from the corresponding authors on reasonable request. Source data are provided with this paper.
Change history
08 October 2021
A Correction to this paper has been published: https://doi.org/10.1038/s43018-021-00283-9
References
Mills, M. N. et al. Management of brain metastases in breast cancer: a review of current practices and emerging treatments. Breast Cancer Res. Treat. 180, 279–300 (2020).
Boire, A., Brastianos, P. K., Garzia, L. & Valiente, M. Brain metastasis. Nat. Rev. Cancer 20, 4–11 (2020).
Kabraji, S. et al. Drug resistance in HER2-positive breast cancer brain metastases: blame the barrier or the brain? Clin. Cancer Res. 24, 1795–1804 (2018).
Arvanitis, C. D., Ferraro, G. B. & Jain, R. K. The blood–brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 20, 26–41 (2019).
Achrol, A. S. et al. Brain metastases. Nat. Rev. Dis. Primers 5, 5 (2019).
Kodack, D. P. et al. The brain microenvironment mediates resistance in luminal breast cancer to PI3K inhibition through HER3 activation. Sci. Transl. Med. 24, 9 (2017).
Monsky, W. L. et al. Role of host microenvironment in angiogenesis and microvascular functions in human breast cancer xenografts: mammary fat pad versus cranial tumors. Clin. Cancer Res. 8, 1008–1013 (2002).
Lyle, L. T. et al. Alterations in pericyte subpopulations are associated with elevated blood–tumor barrier permeability in experimental brain metastasis of breast cancer. Clin. Cancer Res. 22, 5287–5299 (2016).
Gril, B. et al. Reactive astrocytic S1P3 signaling modulates the blood–tumor barrier in brain metastases. Nat. Commun. 9, 2705 (2018).
Zhang, L. et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015).
Valiente, M. et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014).
Chen, Q. et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016).
Priego, N. et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 24, 1024–1035 (2018).
Haddad-Tovolli, R., Dragano, N. R. V., Ramalho, A. F. S. & Velloso, L. A. Development and function of the blood–brain barrier in the context of metabolic control. Front. Neurosci. 11, 224 (2017).
McGale, E. H., Pye, I. F., Stonier, C., Hutchinson, E. C. & Aber, G. M. Studies of the inter-relationship between cerebrospinal fluid and plasma amino acid concentrations in normal individuals. J. Neurochem. 29, 291–297 (1977).
Ngo, B. et al. Limited environmental serine and glycine confer brain metastasis sensitivity to PHGDH inhibition. Cancer Discov. 10, 1352–1373 (2020).
Rainesalo, S. et al. Plasma and cerebrospinal fluid amino acids in epileptic patients. Neurochem. Res. 29, 319–324 (2004).
Dolgodilina, E. et al. Brain interstitial fluid glutamine homeostasis is controlled by blood–brain barrier SLC7A5/LAT1 amino acid transporter. J. Cereb. Blood Flow Metab. 36, 1929–1941 (2016).
Neman, J. et al. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc. Natl Acad. Sci. USA 111, 984–989 (2014).
Zeng, Q. et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 573, 526–531 (2019).
Chi, Y. et al. Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis. Science 369, 276–282 (2020).
Askoxylakis, V. et al. Preclinical efficacy of ado-trastuzumab emtansine in the brain microenvironment. J. Natl Cancer Inst. 108, djv313 (2016).
Baenke, F., Peck, B., Miess, H. & Schulze, A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6, 1353–1363 (2013).
Liu, S., Dai, Z., Cooper, D. E., Kirsch, D. G. & Locasale, J. W. Quantitative analysis of the physiological contributions of glucose to the TCA cycle. Cell Metab. 32, 619–628 (2020).
Mergenthaler, P., Lindauer, U., Dienel, G. A. & Meisel, A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci. 36, 587–597 (2013).
Kamphorst, J. J., Chung, M. K., Fan, J. & Rabinowitz, J. D. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2, 23 (2014).
Buescher, J. M. et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 (2015).
Lane, A. N., Higashi, R. M. & Fan, T. W. Preclinical models for interrogating drug action in human cancers using stable isotope resolved metabolomics (SIRM). Metabolomics 12, 118 (2016).
Sellers, K. et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 125, 687–698 (2015).
Vareslija, D. et al. Transcriptome characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J. Natl Cancer Inst. 111, 388–398 (2019).
Zhang, X. H. et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell 16, 67–78 (2009).
Xu, J. et al. 14-3-3ζ turns TGF-β’s function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell 27, 177–192 (2015).
Rohrig, F. & Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016).
Lewis, C. A. et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 34, 5128–5140 (2015).
Tang, L. & Kebarle, P. Dependence of ion intensity in electrospray mass spectrometry on the concentration of the analytes in the electrosprayed solution. Anal. Chem. 65, 3654–3668 (1993).
Irani, D. Cerebrospinal Fluid in Clinical Practice vol. 1 79 (Elsevier, 2009).
Sullivan, M. R. et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 8, e44235 (2019).
Liu, J. J., Green, P., John Mann, J., Rapoport, S. I. & Sublette, M. E. Pathways of polyunsaturated fatty acid utilization: implications for brain function in neuropsychiatric health and disease. Brain Res. 1597, 220–246 (2015).
Benatti, P., Peluso, G., Nicolai, R. & Calvani, M. Polyunsaturated fatty acids: biochemical, nutritional and epigenetic properties. J. Am. Coll. Nutr. 23, 281–302 (2004).
Liu, Z. et al. Improving orthotopic mouse models of patient-derived breast cancer brain metastases by a modified intracarotid injection method. Sci. Rep. 9, 622 (2019).
Kley, J. T., Mack, J., Hamilton, B., Scheuerer, S. & Redemann, N. Discovery of BI 99179, a potent and selective inhibitor of type I fatty acid synthase with central exposure. Bioorg. Med. Chem. Lett. 21, 5924–5927 (2011).
Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258–272 (2017).
Muir, A. et al. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife 6, e27713 (2017).
Tardito, S. et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 17, 1556–1568 (2015).
Schug, Z. T. et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 (2015).
Vande Voorde, J. et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv. 5, eaau7314 (2019).
Jin, X. et al. MetMap: a map of metastatic potential of human cancer cell lines. Nature 588, 331–336 (2020).
Cordero, A. et al. FABP7 is a key metabolic regulator in HER2+ breast cancer brain metastasis. Oncogene 38, 6445–6460 (2019).
Santana-Codina, N. et al. GRP94 is involved in the lipid phenotype of brain metastatic cells. Int. J. Mol. Sci. 20, 3883 (2019).
Zou, Y. et al. Polyunsaturated fatty acids from astrocytes activate PPARγ signaling in cancer cells to promote brain metastasis. Cancer Discov. 9, 1720–1735 (2019).
Pinkham, K. et al. Stearoyl CoA desaturase is essential for regulation of endoplasmic reticulum homeostasis and tumor growth in glioblastoma cancer stem cells. Stem Cell Rep. 12, 712–727 (2019).
O’Brien, J. S. & Sampson, E. L. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J. Lipid Res. 6, 537–544 (1965).
Sullivan, M. R. et al. Increased serine synthesis provides an advantage for tumors arising in tissues where serine levels are limiting. Cell Metab. 29, 1410–1421 (2019).
Elia, I. et al. Breast cancer cells rely on environmental pyruvate to shape the metastatic niche. Nature 568, 117–121 (2019).
Villa, G. R. et al. An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell 30, 683–693 (2016).
Luengo, A., Gui, D. Y. & Vander Heiden, M. G. Targeting metabolism for cancer therapy. Cell Chem. Biol. 24, 1161–1180 (2017).
Jiang, G. et al. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genomics 17, 525 (2016).
Hosios, A. M., Li, Z., Lien, E. C. & Vander Heiden, M. G. Preparation of lipid-stripped serum for the study of lipid metabolism in cell culture. Bio-protocol 8, e2876 (2018).
Yuan, F. et al. Vascular permeability and microcirculation of gliomas and mammary carcinomas transplanted in rat and mouse cranial windows. Cancer Res. 54, 4564–4568 (1994).
Kodack, D. P. et al. Combined targeting of HER2 and VEGFR2 for effective treatment of HER2-amplified breast cancer brain metastases. Proc. Natl Acad. Sci. USA 109, E3119–E3127 (2012).
Davidson, S. M. et al. Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Vichai, V. & Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 1, 1112–1116 (2006).
Lewis, C. A. et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014).
Heinrich, P. et al. Correcting for natural isotope abundance and tracer impurity in MS-, MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep. 8, 17910 (2018).
Blanc, L., Lenaerts, A., Dartois, V. & Prideaux, B. Visualization of mycobacterial biomarkers and tuberculosis drugs in infected tissue by MALDI-MS imaging. Anal. Chem. 90, 6275–6282 (2018).
Gogolla, N., Galimberti, I., DePaola, V. & Caroni, P. Staining protocol for organotypic hippocampal slice cultures. Nat. Protoc. 1, 2452–2456 (2006).
Lim, N. K. et al. An improved method for collection of cerebrospinal fluid from anesthetized mice. J. Vis. Exp. https://doi.org/10.3791/56774 (2018).
Haslene-Hox, H. et al. A new method for isolation of interstitial fluid from human solid tumors applied to proteomic analysis of ovarian carcinoma tissue. PLoS ONE 6, e19217 (2011).
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016).
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015).
Wiig, H., Aukland, K. & Tenstad, O. Isolation of interstitial fluid from rat mammary tumors by a centrifugation method. Am. J. Physiol. Heart. Circ. Physiol. 284, H416–H424 (2003).
Haug, K. et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48, D440–D444 (2020).
Acknowledgements
We thank the members of the Vander Heiden and Jain Laboratory for helpful discussions. We specifically thank P. Kumar, A. Srinivasan Kumar, J. Willem van Wijnbergen., D. Staiculescu and J. A. Engelman for their input, as well as the MGH and MIT mouse facility and veterinary staff for technical support. We thank H. Lee (MGH Biostatistics) for his helpful input on statistical analysis. We thank Boehringer Ingelheim for providing the BI99179 compound via their opnMe program (https://opnme.com/molecules/fas-bi99179). This work was supported by a Koch Institute/DFHCC Bridge project grant to M.G.V.H. and R.K.J. G.F. received a fellowship from Susan G. Komen for the Cure. A.A. received support as an HHMI Medical Research Fellow. A.L., K.L.A. and S.M.D. were supported by the National Science Foundation and T32GM007287. A.L. was also supported by the Ludwig Center for Molecular Oncology Fund. R.F. was supported by the Novo Nordisk Foundation (NNF10CC1016517) and the Knut and Alice Wallenberg Foundation. L.C.C. acknowledges support from the National Institutes of Health (NIH) (R35CA197588). V.A.D. acknowledges support from shared instrumentation grants (S10OD018072 and S10OD023524). R.K.J. acknowledges support from the NIH (R35CA197742, R01CA208205, U01CA224173), National Foundation for Cancer Research; the Ludwig Center at Harvard; the Jane’s Trust Foundation; the Advanced Medical Research Foundation and by the US Department of Defense Breast Cancer Research Program Innovator Award W81XWH-10-1-0016. M.G.V.H. acknowledges support from a Faculty Scholar grant from the Howard Hughes Medical Institute, Stand Up to Cancer, the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT, the Emerald Foundation and the NIH (R35CA242379, R01CA201276, R01CA168653, P30CA14051).
Author information
Authors and Affiliations
Contributions
G.B.F., A.A. and A.L. performed experiments and helped analyze all data with supervision from R.K.J. and M.G.V.H. D.P.K., D.B. and D.F. contributed to identifying metabolic signatures of brain metastasis; K.L.A. and A.W. contributed to in vitro work and GCMS analysis. A.D. and C.B.C. performed LCMS lipidomics and helped with analysis; L.B., B.P. and V.A.D. performed IMS imaging and analysis; X.J. and T.R.G. contributed to extracellular fluid isolation and provided input on data interpretation. J.M.P., N.I.L. and E.B. collected clinical samples and contributed to analysis of patient tumor sections; J.C. and D.G.D. performed ultrasound imaging of liver tumors; C.R.C. and S.M.D. contributed to in vivo glucose tracing studies. Z.A. performed flow cytometry analysis. R.F. and J.N. helped with analysis of lipidomics data. I.X.C., C.N. and D.E.H. analyzed human expression databases. K.N. performed analysis of Affymetrix array. M.D. and S.R. contributed to CRISPR-Cas9 methodology and animal implantations. C.A.L., L.C.C. and J.M.A. performed targeted metabolomics; G.F., A.A., A.L., R.K.J. and M.G.V.H. designed the study and wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
A.L. is a current employee of a Flagship Pioneering biotechnology start-up company. I.X.C. is a current employee of Stimit Corporation. D.G.D. received consultant fees from Bayer, Simcere and BMS and research grants from Bayer, Exelixis and BMS. L.C.C. is a founder and member of the board of directors of Agios Pharmaceuticals and is a founder and receives research support from Petra Pharmaceuticals. R.K.J received honorarium from Amgen; consultant fees from Chugai, Merck, Ophthotech, Pfizer, SPARC, SynDevRx; owns equity in Accurius, Enlight, Ophthotech, SynDevRx; and serves on the Boards of Trustees of Tekla Healthcare Investors, Tekla Life Sciences Investors, Tekla Healthcare Opportunities Fund, Tekla World Healthcare Fund. Neither any reagent nor any funding from these organizations was used in this study. M.G.V.H. is a scientific advisory board member for Agios Pharmaceuticals, Aeglea Biotherapeutics, Auron Therapeutics, Faeth Therapeutics and iTeos Therapeutics.
Additional information
Peer review information Nature Cancer thanks Almut Schulze and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Histological characterization of MFP and brain BT474 tumors.
a, HER2 immunohistochemistry (IHC) of a BT474 mammary fat pad (MFP) tumor section. b, Immunofluorescence (IF) of ionized calcium binding adaptor molecule 1 (Iba1) and α-smooth muscle actin (SMA) of a consecutive tissue section from the tumor presented in a. c, HER2 IHC of a BT474 brain tumor section. d, IF of Glial fibrillary acidic protein (GFAP) and Iba1 of a consecutive tissue section from the tumor presented in c. For all panels, scale bar = 1 mm.
Extended Data Fig. 2 Assessment of glucose carbon fate in breast tumors growing in different tissue sites.
a, Relative gamma-aminobutyric acid (GABA) levels were measured by liquid chromatography–mass spectrometry (LCMS) in BT474 tumors isolated from the brain or mammary fat pad (MFP) of Nude mice. Data are from the dataset presented in Fig. 1a and Supplementary Data 1. **P = 0.0038 by two-tailed t-test (Brain tumors, n = 7; MFP tumors n = 6; tumors from independent mice). b, Western blot analysis of acetyl-CoA carboxylase (ACC1), fatty acid synthase (FASN), and stearoyl-CoA desaturase-1 (SCD1) expression in MDAMB361 brain and MFP tumor tissue. β-actin expression was assessed as a loading control, and relative densitometry values (normalized to β-actin expression) were used for quantitation with values presented beneath each blot. Differences in protein expression between brain and MFP tumors was compared using a two-tailed t-test. c, The percent of fully labeled glucose (m + 6) in blood plasma following a 12 h 30 mg/kg/min 13C-glucose infusion into Nude female mice bearing BT474 lesions in the brain or in the MFP as assessed by gas chromatography–mass spectrometry (GCMS). **P = 0.0094 by two-tailed t-test. (Plasma from mice bearing BT474 brain tumors, n = 5; Plasma from mice bearing BT474 MFP tumors, n = 6). d-m, The distribution of 13C-labeling in the indicated metabolites as measured by GCMS from BT474 tumors in the brain and MFP, and from noncancerous brain and MFP adipose tissue (WAT) after a 12 h 30 mg/kg/min 13C-glucose infusion into Nude female mice. The data for each isotopologue presented was normalized to 13C-glucose labeling in plasma. (Brain tumors, n = 5; MFP tumors, n = 6; Cortex tissue, n = 12; WAT, n = 5). n,o, BT474 brain and MFP tumors were collected following a 12 h 30 mg/kg/min 13C-glucose infusion and saponified palmitate levels (n) and the distribution of 13C-label in even isotopologues of saponified palmitate (o) were assessed by GCMS. Palmitate levels were normalized to tissue weight and compared using a two-tailed t-test (n.s. denotes not significant). Each isotopologue was normalized to 13C-glucose labeling in plasma and to palmitate total ion counts. These data are from a separate experiment as that shown in Fig. 1e, and were collected to enable normalization based on palmitate total ion counts. (Brain Tumors, n = 3; MFP Tumors, n = 5). Data in panels a, and c–o represent means ± s.e.m.
Extended Data Fig. 3 Higher lipid synthesis in breast cancer lesions when compared to surrounding brain tissue.
a, Negative mode matrix assisted laser desorption/ionization–mass spectrometry imaging (MALDI-MSI) of brain tissue from a separate NSG mouse bearing a BT474 brain tumor than that shown in Fig. 1f,g that had been given 4 daily bolus intraperitoneal injections of 2 g/kg 13C-glucose. The spatial distribution of the indicated isotopologues of palmitate, stearate, oleate, and lysophosphatidylinositol (Lyso PI, 18:0) normalized to total ion counts are shown. b,c, HER2 immunohistochemistry (IHC) staining of brain tissue sections collected from the tumors analyzed by MALDI-MSI in Fig. 1g (b) and in Extended Data Fig. 3a (c). d, Negative mode MALDI-MSI of the m + 2 palmitate isotopologue presented in a normalized to m + 0 palmitate. For all panels, scale bar = 1 mm.
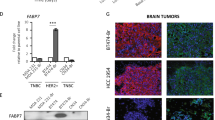
Extended Data Fig. 4 FASN expression is high in breast cancer brain metastases.
a-c, ACC1, ACLY and SCD1 mRNA expression levels from a patient-matched metastatic breast cancer RNAseq dataset30. Matched primary versus brain metastasis (Brain Met.) samples were analyzed separately for the major clinical molecular subtypes. *P = 0.0254 (a), ***P = 0.0003 (b), ****P = 0.0005 (c), by two-tailed paired sample t-test. (Triple negative (TN) tumors, n = 8; Human epidermal growth factor receptor 2 + (HER2 + ) tumors, n = 8; Hormone receptor + (HR + ) tumors, n = 5). d-e, Immunohistochemistry (IHC) and immunofluorescence (IF) assessment of brain metastasis tissue sections derived from 2 different patients with HER2 + breast cancer. IHC was performed to assess HER2 and FASN expression, and IF together with DAPI staining was used to assess glial fibrillary acidic protein (GFAP) and ionized calcium binding adaptor molecule 1 (Iba1) expression on consecutive tissue sections. (scale bar, 1 mm). f, Analysis of SCD1 mRNA expression from a metastatic breast cancer gene expression database comprised of unmatched human tumor samples31,32. Data presented represent mean expression ± s.e.m. *P = 0.0212 (Brain vs Bone), *P = 0.0172 (Brain vs Liver), ***P = 0.0006 by one-way ANOVA followed by Dunnett´s multiple comparisons test (Brain tumors, n = 22; Lung tumors, n = 20; Bone tumors, n = 18; Liver tumors, n = 5).
Extended Data Fig. 5 Impact of extracellular lipids on lipid synthesis enzyme expression and lipid abundance.
a,b, Western blot analysis of FASN, ACC1 and SCD1 in BT474 (a) and MDAMB361 cells (b) cultured in standard (+Lipids) or delipidated (-Lipids) media for 6 d. Lysates were generated from 3 independent samples. β-actin expression was assessed as a loading control. Relative densitometry values (normalized to β-actin expression) were used for quantitation and are presented below each blot. Differences in expression between conditions were compared by a two-tailed t-test. c,d, Relative levels of saponified palmitate measured by GCMS in BT474 (c) and MDAMB361 (d) cells that were cultured in standard (+Lipids) or delipidated (-Lipids) media for 72 h. The samples analyzed are the same as those presented in Fig. 5c. Palmitate levels are normalized to the standard media condition *P = 0.0307 by two-tailed t-test. The data shown represent means ± s.e.m. (n = 3 cell culture biological replicates). e,f, Ratio of complex lipid levels measured by LCMS of BT474 (e) and MDAMB361 (f) cells cultured in standard (+Lipids) or delipidated (-Lipids) media for 6 d. Lipid levels were normalized to protein content as quantified by sulforhodamine b and are presented as a ratio (-/+ Lipids) to show how levels differ based on media lipid availability. A black dotted line indicates a ratio of 1, representing no difference in lipid levels between -Lipids and + Lipids culture conditions. *q < 0.1 by Multiple t-test, False Discovery Rate corrected (n = 3 cell culture biological replicates).
Extended Data Fig. 6 FASN expression is important for breast tumor growth in the brain.
a, Representative ultrasound images used to assess the size of BT474 and MDAMB361 brain tumors (delineated in yellow) in cranial window bearing NSG mice. Summary data is presented in Fig. 5f. (scale bar, 2 mm). b, Western blot analysis of FASN and β-actin expression in control BT474 cells and in BT474 cell clone in which FASN expression is disrupted by CRISPR/Cas9 with sgFASN_2, a different sgRNA than was used to generate the BT474 sgFASN_1 clone presented in Fig. 5a-g. c, d, Growth over time of MFP (c) and brain (d) tumors generated in NSG mice from control or sgFASN_2 BT474 cells. Tumor volumes were measured by caliper or by ultrasound in cranial-window bearing mice, respectively. P values shown were determined using two-way ANOVA (Days × Group). (BT474 control MFP tumors, n = 6; BT474 sgFASN_2 MFP tumors, n = 4; BT474 control brain tumors, n = 7; BT474 sgFASN_2, brain tumors, n = 7). e, Western blot analysis of FASN and β-actin expression in control MDAMB361 lysates or in a MDAMB361 clone in which FASN expression is disrupted CRISPR/Cas9 with sgFASN_2, a different sgRNA than was used to generate the MDAMB361 sgFASN clone presented in Fig. 5a-g. f, Kaplan-Meier survival curve for NSG mice bearing brain tumors produced from control or sgFASN_2 MDAMB361 cells. Median survival was 123 and 190 days for mice bearing MDAMB361 control and sgFASN_2 tumors, respectively. Hazard ratio = 7.842; 95% confidence interval = 1.314 to 46.81. (MDAMB361 control tumors, n = 6; MDAMB361 sgFASN_2 tumors, n = 6). Data in panels c and d represent means ± s.e.m.
Extended Data Fig. 7 FASN expression is important for JIMT1 growth in the brain.
a, Western blot analysis of FASN and β-actin expression in JIMT1 cells in which CRIPSR interference (CRISPRi) was used to disrupt FASN. Cells were transduced with an sgRNA targeting FASN (sgFASN) or a non-targeting control (sgNTC). b, Firefly luciferase-expressing sgNTC or sgFASN JIMT1 cells described in a were implanted into the brains of NSG mice, and tumor burden assessment by whole-body luminescence imaging is show for multiple animals on the indicated days after cell implantation in the brain (n = 6 mice).
Extended Data Fig. 8 FASN knockdown alters cell lipid composition.
a, The indicated acylglycerol levels measured by LCMS from control and FASN-disrupted (sgFASN_1) BT474 tumors growing in the brain of NSG mice are presented as a ratio to show how levels differ based on FASN expression. A black dotted line indicates a ratio of 1, representing no difference in lipid levels between sgFASN_1 and control BT474 tumor tissue. *q < 0.1 by Multiple t-test, False Discovery Rate corrected (n = 4 brain tumors). b, Complex lipid levels measured by LCMS from control and FASN-disrupted (sgFASN_1) BT474 cells in culture (+Lipids) are presented as a ratio to show how levels differ based on FASN expression. A black dotted line indicates a ratio of 1, representing no difference in lipid levels between sgFASN_1 and control cells. Lipid levels were normalized to protein content as determined by sulforhodamine b. All comparisons are significant, q < 0.1 by Multiple t-test, False Discovery Rate corrected. (n = 3 cell culture biological replicates).
Extended Data Fig. 9 FASN is not required for breast tumor growth in the liver.
a, Heatmap showing complex lipid levels measured by LCMS in extracellular fluid (ECF) isolated from the MFP, brain, and liver of non-tumor bearing NSG mice. Data presented within each row were z-score normalized. The data for Brain and MFP ECF are the same as that shown in Fig. 4d. (Brain ECF, n = 2; MFP ECF, n = 2 ECF samples pooled from up to ~8-10 mice). b, Heatmap showing complex lipid levels measured by LCMS of MFP, brain and liver tissue from non-tumor bearing NSG mice. Data presented within each row were z-score normalized. The data for brain and MFP are the same as that shown in Fig. 4e. (Brain tissue, n = 2; MFP tissue, n = 2; Liver Tissue, n = 2). c, Western blot analysis of acetyl-CoA carboxylase (ACC1), ATP citrate lyase (ACLY), fatty acid synthase (FASN), and stearoyl-CoA desaturase-1 (SCD1) in BT474 brain and liver tumor tissue. β-actin expression was assessed as a loading control, and relative densitometry values (normalized to β-actin expression) were used for quantitation and are presented below each blot. Differences in protein expression between brain and liver BT474 lesions was compared by two-tailed t-test. d, The percent of fully labeled (m + 6) glucose in blood plasma following a 12 h 30 mg/kg/min 13C-glucose infusion into female Nude mice bearing BT474 lesions in the liver as assessed GCMS (n = 5 plasma from mice bearing BT474 liver tumors). e, The distribution of pyruvate labeling in BT474 tumors growing in the liver and in non-cancerous liver tissue was measured by GCMS following a 12 h 13C-glucose infusion into female Nude mice. Each isotopologue was normalized to 13C-glucose labeling in plasma (Liver tumor, n = 6; Liver tissue, n = 5). f, The distribution of 13C-label in even isotopologues of palmitate derived from saponified lipids in BT474 liver tumors and in non-cancerous liver tissue following a 12 h 13C-glucose infusion into female Nude mice was measured by GCMS and normalized 13C-glucose labeling in plasma (Liver tumor, n = 6; Liver tissue, n = 5). g, Tumor growth over time of liver tumors in NSG mice generated from control and FASN-disrupted (sgFASN_1) BT474 cells (presented in Fig. 5a-g). Tumor volumes were measured by ultrasound. (BT474 control tumors, n = 4; BT474 sgFASN_1 tumors, n = 4). MAG-monoglyceride; DAG-diglyceride; TAG-triglyceride; Cer-ceramide; LPC- lysophosphatidyl-choline; LPE-lysophosphotidyethanolamine; PC-phosphatidylcholine; PE-phosphatidylethanolamine; PI-phosphatidylinositol; PS-phosphatidylserine; SM-sphingomyelin; CE-cholesteryl ester; @-sphingosine, palmitoylethanolamide, 7−Dehydrodesmosterol, cholesterol, coenzyme Q9, coenzyme Q10. Data presented in panels d, e, f and g are means ± s.e.m.
Extended Data Fig. 10 Lipid availability determines response to pharmacological FASN inhibition.
a-c, BT474 brain tumor-derived organotypic slice cultures were treated for 6 d with vehicle or 500 nM TVB3166 in standard (+Lipids) or delipidated (-Lipids) media. The percent of apoptotic and viable HER2 positive BT474 cells was determined by measuring Annexin V (AV) and 7-amino actinomycin D (7AAD) uptake by flow cytometry. The flow cytometry gating strategy is presented in Supplementary Fig. 2. *P = 0.0111, ***P = 0.0003 by one-way ANOVA followed by Tukey´s test (n = 5 BT474 brain tumor slices). d,e, Mouse weights for the cohort presented in Fig. 6f,g that was treated daily with BI99179 (15 mg/kg) or vehicle control by oral gavage (n = 5 mice). Data presented in all panels are means ± s.e.m.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables 1–3
Supplementary Table 1: Gene array analysis. Supplementary Table 2: gRNA and oligonucleotide sequences. Supplementary Table 3: Commercial reagent information.
Supplementary Data 1
Metabolomics data. Used in Fig. 1.
Supplementary Data 2
Lipidomics data. Used in Fig. 4 and Extended Data Figs. 5, 8 and 9.
Supplementary Data 3
Raw data used for statistical analyses. Used in Figs. 3, 5 and 6 and Extended Data Figs. 2, 4–6 and 10
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots
Source Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Ferraro, G.B., Ali, A., Luengo, A. et al. Fatty acid synthesis is required for breast cancer brain metastasis. Nat Cancer 2, 414–428 (2021). https://doi.org/10.1038/s43018-021-00183-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00183-y
This article is cited by
-
Recent advances in the potential role of RNA N4-acetylcytidine in cancer progression
Cell Communication and Signaling (2024)
-
Loss of the extracellular matrix glycoprotein EMILIN1 accelerates Δ16HER2-driven breast cancer initiation in mice
npj Breast Cancer (2024)
-
Redox signalling regulates breast cancer metastasis via phenotypic and metabolic reprogramming due to p63 activation by HIF1α
British Journal of Cancer (2024)
-
The implications of FASN in immune cell biology and related diseases
Cell Death & Disease (2024)
-
Metabolic heterogeneity in cancer
Nature Metabolism (2024)