Abstract
Cyclin-dependent kinases (CDKs) 4 and 6 inhibitors (CDK4/6is) are effective in metastatic breast cancer, but they have been only modestly effective in most other tumor types. Here we show that tumors expressing low CDK6 rely on CDK4 function and are exquisitely sensitive to CDK4/6is. In contrast, tumor cells expressing both CDK4 and CDK6 have increased reliance on CDK6 to ensure cell cycle progression. We discovered that CDK4/6is and CDK4/6 degraders potently bind and inhibit CDK6 selectively in tumors in which CDK6 is highly thermo-unstable and strongly associated with the HSP90–CDC37 complex. In contrast, CDK4/6is and CDK4/6 degraders are ineffective in antagonizing tumor cells expressing thermostable CDK6, due to their weaker binding to CDK6 in these cells. Thus, we uncover a general mechanism of intrinsic resistance to CDK4/6is and CDK4/6i-derived degraders and the need for new inhibitors targeting the CDK4/6i-resistant, thermostable form of CDK6 for application as cancer therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
MS raw files for CDK6 complex analysis are available at the Mass Spectrometry Interactive Virtual Environment (https://massive.ucsd.edu) under ID MSV000086571.
MS files for global protein degradation have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023137.
Normalized protein quantification data and cell line omics data can be downloaded at DepMap (https://depmap.org/portal/).
Reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application. Source data are provided with this paper.
Code availability
Data analysis was performed in R v.3.6.3 using custom-made or publicly available R packages. The code is available from GitHub (https://github.com/lijin0303/CDK46_expression_dependency). Figure 3e and Extended Data Fig. 3b were generated with R 3.6.3 (Packages taigr, dplyr, tidyr, ggplot2, ggpubr) and corresponding code can be found in https://github.com/lijin0303/CDK46_expression_dependency. Source data are provided with this paper.
References
Sherr, C. J., Beach, D. & Shapiro, G. I. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 6, 353–367 (2016).
O’Leary, B., Finn, R. S. & Turner, N. C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13, 417–430 (2016).
Otto, T. & Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 17, 93–115 (2017).
Finn, R. S. et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 375, 1925–1936 (2016).
Hortobagyi, G. N. et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N. Engl. J. Med. 375, 1738–1748 (2016).
Ingham, M. & Schwartz, G. K. Cell-cycle therapeutics come of age. J. Clin. Oncol. https://doi.org/10.1200/JCO.2016.69.0032 (2017).
Goetz, M. P. et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol. 35, 3638–3646 (2017).
Im, S. A. et al. Overall survival with ribociclib plus endocrine therapy in breast cancer. N. Engl. J. Med. 381, 307–316 (2019).
Knudsen, E. S. & Witkiewicz, A. K. The strange case of CDK4/6 Inhibitors: mechanisms, resistance, and combination strategies. Trends Cancer 3, 39–55 (2017).
Hamilton, E. & Infante, J. R. Targeting CDK4/6 in patients with cancer. Cancer Treat. Rev. 45, 129–138 (2016).
Gong, X. et al. Genomic aberrations that activate D-type cyclins are associated with enhanced sensitivity to the CDK4 and CDK6 inhibitor abemaciclib. Cancer Cell 32, 761–776 (2017).
Kim, S. et al. The potent and selective cyclin-dependent kinases 4 and 6 inhibitor ribociclib (LEE011) is a versatile combination partner in preclinical cancer models. Oncotarget 9, 35226–35240 (2018).
Yang, C. et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene https://doi.org/10.1038/onc.2016.379 (2016).
Placke, T. et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 124, 13–23 (2014).
Taipale, M. et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 (2012).
Vaughan, C. K. et al. Structure of an Hsp90-Cdc37-CDK4 complex. Mol. Cell 23, 697–707 (2006).
Stepanova, L., Leng, X., Parker, S. B. & Harper, J. W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes CDK4. Genes Dev. 10, 1491–1502 (1996).
Taipale, M. et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat. Biotechnol. 31, 630–637 (2013).
Grbovic, O. M. et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc. Natl Acad. Sci. USA 103, 57–62 (2006).
Sawai, A. et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res. 68, 589–596 (2008).
Hallett, S. T. et al. Differential regulation of G1 CDK complexes by the Hsp90-Cdc37 chaperone system. Cell Rep. 21, 1386–1398 (2017).
Lai, A. C. & Crews, C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017).
Brand, M. et al. Homolog-selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem. Biol. 26, 300–306 e309 (2019).
Jiang, B. et al. Development of dual and selective degraders of cyclin-dependent kinases 4 and 6. Angew. Chem. Int. Ed. Engl. 58, 6321–6326 (2019).
Ruscetti, M. et al. NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422 (2018).
Finn, R. S. et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11, R77 (2009).
Goldman, J. W. et al. A randomized phase 3 study of abemaciclib versus erlotinib in previously treated patients with stage IV NSCLC with KRAS mutation: JUNIPER. J. Clin. Oncol. 36, 9025 (2018).
Pacheco, J. & Schenk, E. CDK4/6 inhibition alone and in combination for non-small cell lung cancer. Oncotarget 10, 618–619 (2019).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Kato, J. Y., Matsuoka, M., Strom, D. K. & Sherr, C. J. Regulation of cyclin D-dependent kinase 4 (CDK4) by CDK4-activating kinase. Mol. Cell. Biol. 14, 2713–2721 (1994).
McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
McDonald, E. R. 3rd et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592 (2017).
Nusinow, D. P. et al. Quantitative proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402 (2020).
Marcotte, R. et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 164, 293–309 (2016).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Chen, P. et al. Spectrum and degree of CDK drug interactions predicts clinical performance. Mol. Cancer Ther. 15, 2273–2281 (2016).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 11, 611–617 (2015).
Winter, G. E. et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Chamberlain, P. P. et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 21, 803–809 (2014).
Zhang, C. et al. Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase (ALK). Eur. J. Med. Chem. 151, 304–314 (2018).
Fischer, E. S. et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
Lebraud, H., Wright, D. J., Johnson, C. N. & Heightman, T. D. Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. ACS Cent. Sci. 2, 927–934 (2016).
Sumi, N. J., Kuenzi, B. M., Knezevic, C. E., Remsing Rix, L. L. & Rix, U. Chemoproteomics reveals novel protein and lipid kinase targets of clinical CDK4/6 inhibitors in lung cancer. ACS Chem. Biol. 10, 2680–2686 (2015).
Bisi, J. E. et al. Preclinical development of G1T38: a novel, potent and selective inhibitor of cyclin dependent kinases 4/6 for use as an oral antineoplastic in patients with CDK4/6 sensitive tumors. Oncotarget 8, 42343–42358 (2017).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 (2014).
Patricelli, M. P. et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 18, 699–710 (2011).
Verba, K. A. et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547 (2016).
Boczek, E. E. et al. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc. Natl Acad. Sci. USA 112, E3189–E3198 (2015).
Zhang, J., Yang, P. L. & Gray, N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Bockstaele, L., Bisteau, X., Paternot, S. & Roger, P. P. Differential regulation of cyclin-dependent kinase 4 (CDK4) and CDK6, evidence that CDK4 might not be activated by CDK7, and design of a CDK6 activating mutation. Mol. Cell. Biol. 29, 4188–4200 (2009).
Nomanbhoy, T. K. et al. Chemoproteomic evaluation of target engagement by the cyclin-dependent kinase 4 and 6 inhibitor palbociclib correlates with cancer cell response. Biochemistry 55, 5434–5441 (2016).
Romano, G. et al. A preexisting rare PIK3CA(E545K) subpopulation confers clinical resistance to MEK plus CDK4/6 inhibition in NRAS melanoma and is dependent on S6K1 signaling. Cancer Discov. 8, 556–567 (2018).
Herrera-Abreu, M. T. et al. Early adaptation and acquired resistance to CDK4/6 inhibition in estrogen receptor-positive breast cancer. Cancer Res. 76, 2301–2313 (2016).
Costa, C. et al. PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kα inhibitors in breast cancer. Cancer Discov. 10, 72–85 (2020).
de Leeuw, R. et al. MAPK reliance via acquired CDK4/6 inhibitor resistance in cancer. Clin. Cancer Res. 24, 4201–4214 (2018).
Li, Z. et al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the hippo pathway. Cancer Cell 34, 893–905 (2018).
Guiley, K. Z. et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science https://doi.org/10.1126/science.aaw2106 (2019).
Green, J. L. et al. Direct CDKN2 modulation of CDK4 alters target engagement of CDK4 inhibitor drugs. Mol. Cancer Ther. 18, 771–779 (2019).
Nusinow P. & Gygi, S. P. A guide to the quantitative proteomic profiles of the Cancer Cell Line Encyclopedia. Preprint at bioRxiv https://doi.org/10.1101/2020.02.03.932384 (2020).
Acknowledgements
We thank A. Lujambio, S. Parekh, R. Parsons, E.P. Reddy, R. Sachidanandam, (Icahn School of Medicine at Mount Sinai), I. Aifantis (New York University), S.M. Buckley (University of Nebraska), C.A. Pratilas (Johns Hopkins University) and W.G. Kaelin Jr. (Harvard Medical School) for sharing cell lines and reagents. P.I.P. is supported by the US National Institutes of Health (NIH)/NCI (R01CA204314, R01 CA240362 and R01CA238229), the Irma T. Hirschl Trust, the Manhasset Women’s Coalition against Breast Cancer, the Breast Cancer Alliance, the Melanoma Research Foundation, the Melanoma Research Alliance and Tisch Cancer Institute developmental awards. T.A.A. has been supported by grant T32CA078207 and Z.K. would like to acknowledge the 2017 Robin Chemers Neustein Postdoctoral Fellowship. W.R.S. acknowledges support from the NIH (1R01CA233626) and the Ludwig Center at Harvard, T.I. is supported by the Department of Defense Peer Reviewed Cancer Research Program Horizon Award (W81XWH-19-1-0271). X.C. is supported by the NIH (R01-GM133107). S.A.A. acknowledges support from a grant from the Breast Cancer Research Foundation. This work was supported in part by the P30CA196521 grant (to J.J.) from the NIH and an endowed professorship from the Icahn School of Medicine at Mount Sinai (to J.J.). This work utilized the AVANCE NEO 600 MHz NMR Spectrometer System that was upgraded with funding from an NIH SIG grant 1S10OD025132-01A1.
Author information
Authors and Affiliations
Contributions
X.W. conceived of the study, performed most of the experiments, analyzed the data and helped write the manuscript. X.Y. and Y.X. synthesized chemical compounds. T.A.A, Z.K. and C.A. performed experiments and analyzed data. S.A.A designed research, analyzed data and helped write the manuscript. J.L. and J.J. designed chemical compounds and analyzed data. L.X. carried out MS analysis. B.U. and X.C. supervised MS analysis. R.L., L.W., T.I. and H.W. performed data analysis. S.G.B. and W.R.S. designed the research, analyzed the data and helped to write the manuscript. P.I.P conceived of the study, designed research, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
X.W., X.Y., Y.X., T.A., Z.K., J.L., J.J. and P.I.P. hold a patent on CDK4/6-directed degradation (WO/2018/106870). The Jian Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics and Cullgen, Inc. J.J. is an equity shareholder and consultant of Cullgen, Inc. H.W. and S.G.B. are employees of Eli Lilly. All other authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks John Brognard, Keiran Smalley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
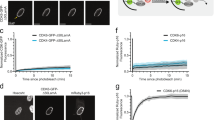
Extended Data Fig. 1 Intrinsic resistance to CDK4/6i is associated with incomplete inhibition of Rb/E2F and expression of CDK6.
a, Cell growth crystal violet assay for the indicated cell lines treated with increasing concentrations of PB for 10-16 days and stained with crystal violet. CDK4/6i-sensitive cell lines were highlighted in blue, CDK4/6i-resistant cell lines were in red. b, MCF7 and HCT116 were treated with 1 µM PB for 24, 48 and 72 hr and lysates were immunoblotted with the indicated antibodies. c, Colo205 cells were treated with 1 µM PB at the indicated time points. Cell lysates were immunoblotted with the indicated antibodies.
Extended Data Fig. 2 Low expression of CDK6 predicts for sensitivity to CDK4/6i in NSCLC.
a, The indicated cell lines were treated with increasing concentrations of PB for 24 hr. Lysates were immunoblotted with the indicated antibodies. b, GI90 values of PB and CDK4/6 dependency in NSCLC cell lines.
Extended Data Fig. 3 Tumors expressing both CDK4 and CDK6 depend selectively on CDK6.
a, A673 and TC-71 cells were transfected with non-targeting control or siCDK4 or siCDK6 for 72 hr. Lysates were immunoblotted with the indicated antibodies. b, Relationship between CDK4 and CDK6 expression (CCLE RNA-seq) and DepMap CRISPR–Cas9 single-gene knockout scores (CERES; 20Q1 public dataset). All expression values are in log2(TPM +1). Cell lines harboring COSMIC hotspot mutations to RB1 are annotated in orange. P-values were calculated based on linear regression analysis.
Extended Data Fig. 4 Development of MS140, a potent and selective CDK4/6-degrader (PROTAC).
a. IC50 of in vitro kinase activity assays for PB and MS140 against CDK4/cyclin D1 and CDK6/cyclin D1. b, T47D cells were pretreated with either the proteasome inhibitor 100 nM bortezomib (BOR), 10 µM PB, 10 µM pomalidomide (POM) or 1 µM MLN4924 (MLN) for 4 hr, followed by treatment with MS140 (100 nM/3 hr). Lysates were subjected to immunoblotting with the indicated antibodies. c, Chemical structure of the MS140 negative control (MS140-ve) that does not bind CRBN.
Extended Data Fig. 5 CDK4/6-directed degradation is more effective than CDK4/6i in CDK4/6i-S tumor cells.
a, MCL cell lines were treated with 0.1 µM PB or MS140 at different time points. Lysates were immunoblotted with the indicated antibodies. b, Colo205 cells expressing Dox-inducible shCDK4 or shCDK6 were treated with or without 0.1 µg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies. c, Colo205 cells expressing Dox-inducible shCDK4 or shCDK6 were treated with or without 0.1 µg/ml doxycycline for 10 days followed by crystal violet staining. d, Dependency score of CDK4 and CDK6 from cancer cell line encyclopedia (CCLE) and Depmap portal database. e, GI50 values of PB and MS140 in hematologic cancer cell lines. f, Growth curve for an efficacy assay in JeKo-1 tumor xenografts in nude mice treated with vehicle or MS140 (25 mg/kg, b.i.d) or PB (50 mg/kg, q.d.) for 21 days. Each treatment contained 8 animals (n=8). Data represent mean ± SEM. g, Body weight in mice bearing JeKo-1 tumors treated with vehicle (n=8) or PB (50 mg/kg, q.d., n=8) or MS140 (25 mg/kg, b.i.d., n=8) in the course of the experiment (21 days). Data are presented as mean ± S.D. h, White blood cell, lymphocytes and red blood cell counts in C57BL/6 mice before treatment and post treatment with PB (50 mg/kg, q.d., n=8) or MS140 (25 mg/kg, b.i.d., n=7) for 21 days. Data are presented as mean ± S.D. Statistical significance was determined by paired two-tailed Student’s t-test.
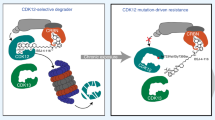
Extended Data Fig. 6 In CDK4/6-R cells, CDK4/6 degraders fail to degrade CDK6 due to weak binding of compound.
a, Calu6 cells transiently expressing pcDNA3 (Ev) or pcDNA3-Flag-CRBN were treated with increasing concentrations of MS140 for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. b, MV4-11 and A375 were treated MS140 (3 nM) or YKL-06-102 (3 nM) or BSJ-02-162 (3 nM) at different time points. Lysates were subjected to immunoblotting with the indicated antibodies. c, KMS-12-PE and Calu6 were treated MS140 (3 nM) or YKL-06-102 (3 nM) or BSJ-02-162 (3 nM) at different time points. Lysates were subjected to immunoblotting with the indicated antibodies.
Extended Data Fig. 7 CDK4/6i-resistant cells express CDK6 as a thermostable, weak HSP90 client protein.
a, Comparison of total peptide-spectrum match (PSM) for CDK6-interacting proteins by mass spectrometry in KMS-12-PE and Calu6. b, Cell lysates from Colo205 and Calu6 were either subjected to Co-IP with a CDK6 antibody followed by immunoblotting with HSP90, CDC37 and CDK6, or immunoblotted with the indicated antibodies. c, The indicated cell lines were treated with increasing concentrations of Ganetespib (GAN) for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. d, The indicated cell lines were treated with 40 nM Luminespib (LUM) at the indicated time points. Lysates were subjected to immunoblotting with the indicated antibodies. e. Calu6 cell line expressing Dox-inducible shCDC37 were treated with or without 0.1 µg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies. f, CDK4-dependent cell lines were treated with 30 nM GAN for the indicated time points. Lysates were subjected to immunoblotting with the indicated antibodies. g, KMS-12-PE and Calu6 were treated with increasing concentrations of GAN for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. h, Lysates from A375 cells ectopically expressing V5-CDK6 or V5-CDK6 S178p were immunoprecipited with a V5 antibody. The immunoprecipitates were subjected to kinase assay with recombinant Rb protein as substrate.
Supplementary information
Supplementary Table 1
Oligonucleotide sequences
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Wu, X., Yang, X., Xiong, Y. et al. Distinct CDK6 complexes determine tumor cell response to CDK4/6 inhibitors and degraders. Nat Cancer 2, 429–443 (2021). https://doi.org/10.1038/s43018-021-00174-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00174-z
This article is cited by
-
ERK hyperactivation serves as a unified mechanism of escape in intrinsic and acquired CDK4/6 inhibitor resistance in acral lentiginous melanoma
Oncogene (2024)
-
PROTAC-mediated CDK degradation differentially impacts cancer cell cycles due to heterogeneity in kinase dependencies
British Journal of Cancer (2023)
-
The RNA-binding protein LRPPRC promotes resistance to CDK4/6 inhibition in lung cancer
Nature Communications (2023)
-
LIMK2 promotes melanoma tumor growth and metastasis through G3BP1-ESM1 pathway-mediated apoptosis inhibition
Oncogene (2023)
-
Cellular mechanisms underlying response and resistance to CDK4/6 inhibitors in the treatment of hormone receptor-positive breast cancer
Breast Cancer Research (2022)