Abstract
Argininosuccinate synthase (ASS1) downregulation in different tumors has been shown to support cell proliferation and yet, in several common cancer subsets ASS1 expression associates with poor patient prognosis. Here we demonstrate that ASS1 expression under glucose deprivation is induced by c-MYC, providing survival benefit by increasing nitric oxide synthesis and activating the gluconeogenic enzymes pyruvate carboxylase and phosphoenolpyruvate carboxykinase by S-nitrosylation. The resulting increased flux through gluconeogenesis enhances serine, glycine and subsequently purine synthesis. Notably, high ASS1-expressing breast cancer mice do not respond to immune checkpoint inhibitors and patients with breast cancer with high ASS1 have more metastases. We further find that inhibiting purine synthesis increases pyrimidine to purine ratio, elevates expression of the immunoproteasome and significantly enhances the response of autologous primary CD8+ T cells to anti-PD-1. These results suggest that treating patients with high-ASS1 cancers with purine synthesis inhibition is beneficial and may also sensitize them to immune checkpoint inhibition therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Whole-exome sequencing data that support the findings of this study have been deposited in the SRA repository under accession code PRJNA637021. Transcriptomics, whole-exome sequencing and clinical data for Figs. 1, 4 and 5 from the TCGA Research Network: http://cancergenome.nih.gov/. The transcriptomics and clinical data derived from this resource that supports the findings of this study are available in the UCSC Xena browser (https://xena.ucsc.edu): https://toil.xenahubs.net/download/TcgaTargetGtex_RSEM_Hugo_norm_count.gz and https://toil.xenahubs.net/download/TcgaTargetGTEX_phenotype.txt.gz. Whole-exome sequencing data were downloaded from cBioPortal (https://www.cbioportal.org/). CPTAC proteomics data (https://proteomics.cancer.gov/programs/cptac) that were re-analyzed here are available under accession code S046. Source data for Figs. 1–6 and Extended Data Figs. 1–5 are provided as Source Data files. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Code availability
The custom code used for TCGA analysis in the revised manuscript is available via the GitHub repository at https://www.github.com/jooslee/ASS1. Source data are provided with this paper.
References
Husson, A., Brasse-Lagnel, C., Fairand, A., Renouf, S. & Lavoinne, A. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur. J. Biochem. 270, 1887–1899 (2003).
Erez, A., Nagamani, S. C. & Lee, B. Argininosuccinate lyase deficiency-argininosuccinic aciduria and beyond. Am. J. Med. Genet. 157, 45–53 (2011).
Liu, Q. et al. Reduced expression of argininosuccinate synthetase 1 has a negative prognostic impact in patients with pancreatic ductal adenocarcinoma. PLoS ONE 12, e0171985 (2017).
Sahu, D. et al. Argininosuccinate synthetase 1 loss in invasive bladder cancer regulates survival through general control nonderepressible 2 kinase-mediated eukaryotic initiation factor 2α activity and is targetable by pegylated arginine deiminase. Am. J. Pathol. https://doi.org/10.1016/j.ajpath.2016.09.004 (2016).
Kim, Y. et al. Reduced argininosuccinate synthetase expression in refractory sarcomas: impacts on therapeutic potential and drug resistance. Oncotarget 7, 70832–70844 (2016).
Wu, L. et al. Expression of argininosuccinate synthetase in patients with hepatocellular carcinoma. J. Gastroenterol. Hepatol. 28, 365–368 (2013).
Rabinovich, S. et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383 (2015).
Kim, J. et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172 (2017).
Lee, J. S. et al. Urea cycle dysregulation generates clinically relevant genomic and biochemical signatures. Cell 174, e1522 (2018).
Delage, B. et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 126, 2762–2772 (2010).
Szlosarek, P. W. et al. Aberrant regulation of argininosuccinate synthetase by TNF-α in human epithelial ovarian cancer. Int. J. Cancer 121, 6–11 (2007).
Vincent, E. E. et al. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 (2015).
Mihaylova, M. M. et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 (2011).
Mesonero, J., Matosin, M., Cambier, D., Rodriguez-Yoldi, M. J. & Brot-Laroche, E. Sugar-dependent expression of the fructose transporter GLUT5 in Caco-2 cells. Biochem. J. 312(Pt 3), 757–762 (1995).
Bean, G. R. et al. A metabolic synthetic lethal strategy with arginine deprivation and chloroquine leads to cell death in ASS1-deficient sarcomas. Cell Death Dis. 7, e2406 (2016).
Bateman, L. A. et al. Argininosuccinate synthase 1 is a metabolic regulator of colorectal cancer pathogenicity. ACS Chem. Biol. 12, 905–911 (2017).
Miles, L., Heubi, J. E. & Bove, K. E. Hepatocyte glycogen accumulation in patients undergoing dietary management of urea cycle defects mimics storage disease. J. Pediatr. Gastr. Nutr. 40, 471–476 (2005).
Kuhara, H. et al. Neonatal type of argininosuccinate synthetase deficiency. Report of two cases with autopsy findings. Acta Pathol. Jpn. 35, 995–1006 (1985).
Bigot, A., Tchan, M. C., Thoreau, B., Blasco, H. & Maillot, F. Liver involvement in urea cycle disorders: a review of the literature. J. Inherit. Metab. Dis. 40, 757–769 (2017).
Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999).
Perez, C. J. et al. Two hypomorphic alleles of mouse ass1 as a new animal model of citrullinemia type I and other hyperammonemic syndromes. Am. J. Pathol. 177, 1958–1968 (2010).
Pennisi, P. et al. Recombinant human insulin-like growth factor-I treatment inhibits gluconeogenesis in a transgenic mouse model of type 2 diabetes mellitus. Endocrinology 147, 2619–2630 (2006).
Rashed, H. M., Nair, B. G. & Patel, T. B. Regulation of hepatic glycolysis and gluconeogenesis by atrial natriuretic peptide. Arch. Biochem. Biophys. 298, 640–645 (1992).
Zareba-Koziol, M., Szwajda, A., Dadlez, M., Wyslouch-Cieszynska, A. & Lalowski, M. Global analysis of S-nitrosylation sites in the wild type (APP) transgenic mouse brain-clues for synaptic pathology. Mol. Cell Proteomics 13, 2288–2305 (2014).
Pae, H. O. et al. Nitric oxide protects murine embryonic liver cells (BNL CL.2) from cytotoxicity induced by glucose deprivation. Pharmacol. Toxicol. 86, 140–144 (2000).
Cherry, P. D. & Wolin, M. S. Ascorbate activates soluble guanylate cyclase via H2O2-metabolism by catalase. Free Radic. Biol. Med. 7, 485–490 (1989).
Keilhoff, G., Reiser, M., Stanarius, A., Aoki, E. & Wolf, G. Citrulline immunohistochemistry for demonstration of NOS activity in vivo and in vitro. Nitric Oxide 4, 343–353 (2000).
Park, S. W. et al. Nitric oxide upregulates the cyclooxygenase-2 expression through the cAMP-response element in its promoter in several cancer cell lines. Oncogene 24, 6689–6698 (2005).
Sun, L. et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 25, 429–444 (2015).
Tsai, W. B. et al. Resistance to arginine deiminase treatment in melanoma cells is associated with induced argininosuccinate synthetase expression involving c-Myc/HIF-1α/Sp4. Mol. Cancer Ther. 8, 3223–3233 (2009).
Follis, A. V., Hammoudeh, D. I., Wang, H., Prochownik, E. V. & Metallo, S. J. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem. Biol. 15, 1149–1155 (2008).
Erez, A. et al. Requirement of argininosuccinate lyase for systemic nitric oxide production. Nat. Med. 17, 1619–1626 (2011).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
Burrows, N. et al. Hypoxia-induced nitric oxide production and tumour perfusion is inhibited by pegylated arginine deiminase (ADI-PEG20). Sci. Rep. 6, 22950 (2016).
Yokota, S. Mizoribine: mode of action and effects in clinical use. Pediatr. Int. 44, 196–198 (2002).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Lee, J. S. et al. Whole exome precision oncology targeting synthetic lethal vulnerabilities across the tumor transcriptome. Preprint at bioRxiv https://doi.org/10.1101/2020.02.16.951699 (2020).
Spits, M. & Neefjes, J. Immunoproteasomes and immunotherapy-a smoking gun for lung cancer? J. Thorac. Dis. 8, E558–E563 (2016).
Basler, M., Kirk, C. J. & Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 25, 74–80 (2013).
Lee, M. et al. Expression of immunoproteasome subunit LMP7 in breast cancer and its association with immune-related markers. Cancer Res. Treat. 51, 80–89 (2019).
Vijayan, D., Young, A., Teng, M. W. L. & Smyth, M. J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 17, 709–724 (2017).
Fu, Y., Liu, S., Zeng, S. & Shen, H. The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol. Cancer 17, 62 (2018).
Shan, Y. S. et al. Argininosuccinate synthetase 1 suppression and arginine restriction inhibit cell migration in gastric cancer cell lines. Sci. Rep. 5, 9783 (2015).
Cheng, S. H. et al. Validation of the 18-gene classifier as a prognostic biomarker of distant metastasis in breast cancer. PLoS ONE 12, e0184372 (2017).
Montal, E. D. et al. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol. Cell 60, 571–583 (2015).
Possemato, R. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011).
Antonov, A. et al. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget 5, 11004–11013 (2014).
DeBerardinis, R. J. & Chandel, N. S. Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016).
Erb, N., Harms, D. O. & Janka-Schaub, G. Pharmacokinetics and metabolism of thiopurines in children with acute lymphoblastic leukemia receiving 6-thioguanine versus 6-mercaptopurine. Cancer Chemoth. Pharm. 42, 266–272 (1998).
Forrester, M. T. et al. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 (2009).
Stettner, N. et al. Induction of nitric-oxide metabolism in enterocytes alleviates colitis and inflammation-associated colon cancer. Cell Rep. 23, 1962–1976 (2018).
Goldman, M. et al. The UCSC Xena Platform for cancer genomics data visualization and interpretation. Preprint at bioRxiv https://doi.org/10.1101/326470 (2018).
Mantel, N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 50, 163–170 (1966).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Janin, J. Surface and inside volumes in globular proteins. Nature 277, 491–492 (1979).
Cheng, S. H. et al. Validation of the 18-gene classifier as a prognostic biomarker of distant metastasis in breast cancer. PLoS ONE 12, e0184372 (2017).
Acknowledgements
We acknowledge and thank the Weizmann Institute for providing financial and infrastructural support. We thank S. Mcguirck, McGill University, Montreal, Canada, for assistance with GC–MS data analysis. The MC-38 cancer cell line was kindly given to us by E. Elinav from the Weizmann Institute of Science. A.E. is incumbent of the Leah Omenn Career Development Chair and is supported by research grants from the European research program (ERC818943) and from the Israel Science Foundation (860/18). A.E. received additional support from The Moross Integrated Cancer Center, Sagol Institute for Longevity Research, Adelis Foundation, Rising Tide Foundation and from Manya and Adolph Zarovinsky. L.A. receives support from Robin Lynn & Lawrence S. Blumberg. E. Ruppin gratefully acknowledges support from NCI/CCR intramural funding. J.S.L. is partly supported by a grant of the National Research Foundation of Korea funded by the Korean Government (NRF-2020R1A2C2007652). A.P. is supported by the NRF-ISF grant 3127/19.
Author information
Authors and Affiliations
Contributions
R. Keshet, J.S.L. and L.A. led the genomic analysis and molecular experiments; Y.A., L.Q.J.L., S.L., S.R., H.W.T., H.L. and S.G., helped with the molecular experiments; R.O., N.S., Y.K., Y. Kuznetsov and O.G. were responsible for the animal experiments; M.T., S.M., A.B., M. Itkin and T.M. contributed to the metabolomic analysis; R. Katzir, Y.Z., K.T., T.W.S. and I.U. helped with the genomic analysis. M. Iraqi, A.P. and N.P. performed the PDX experiments. S.E.K. kindly provided the ASS1 flox mice. E.R. and A.E. initiated and led the study and wrote the manuscript. All authors were involved in discussions about study design, contributed to the writing and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
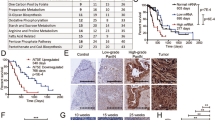
Extended Data Fig. 1 ASS1 expression supports cancer survival under glucose deprivation.
a, The 10 cancer types with high ASS1 (grey, n = 5672; Fig. 1a left panel) demonstrate a significantly lower UCD-score as compared to 10 cancer types with low ASS1 expression (white, n = 6666) following Fig. 1a; (one-sided Wilcoxon ranksum P < 2.2E-16). The box is drawn from top quartile (75-percentile) to bottom quartile (25-percentile) of the data with a horizontal line drawn in the middle to denote the median value. The lowest point of the bottom whisker is the minimum of the data and the highest point of the top whisker is the maximum of the data. Right panel- The figure shows the Spearman rank correlation coefficients (Y-axis) between UCD-score and the expression of each of UC enzymes across TCGA samples. b, Western blot (left) and band intensity quantification relative to GAPDH (right) showing elevation in ASS1, PC and PCK in A549 grown in glucose deficient medium for 48 h. c, Top panel - 4T1 and SW620 cells were grown either in normal medium or in medium without glucose for 48 h. ASS1 levels were analyzed by western blotting. Bottom panel - A western blot showing ASS1 levels in MNNG human osteosarcoma cancer cells grown either in normal medium (control), without serum (NS) or without glucose (NG). d, ASS1 supports glucose-independent survival in murine breast cancer cells and in human colon cancer cells. 4T1 and SW620 cells (expressing either ASS1-shRNA or control GFP-shRNA) were grown either with or without glucose for 48 h (4T1 cells) or 96 h (SW620 cells). Survival was measured by crystal violet staining. Results were normalized to ASS1 expression levels (left panels) or to survival ratio of control cells in glucose-free medium compared to glucose-full medium. n = 3 biological replicates in each group in 4T1 gene expression; n = 3–4 biological replicates in each group in 4T1 survival assay as depicted in the graph; n = 2 biological replicates in each group in SW620 gene expression. In SW620 survival assay presented are technical replicates of a single representative experiment – mean shASS1#1 survival is 30% of control and for shASS1#2 is 74% of control. p value was calculated using two-tailed student’s t-test. In 4T1 gene expression p = 8.8E-07 for control vs. shASS1#1; p = 1.8E-06 for control vs. shASS1#2. In 4T1 survival assay p = 0.032 for control vs.shASS1#1; p = 0.048 for control vs. shASS1#2. In SW620 gene expression p = 0.007 for control vs. shASS1#1; p = 0.006 for control vs. shASS1#2. e, Left panel- RT PCR for ASS1 mRNA levels in A549 control cells versus shASS and shASS1 with ASS1 over expression (OE). Right panel- FACS annexin –shASS1 increases apoptosis. A549 cells were grown in glucose and serum free medium for 72 hrs. Annexin V/7ADD assay was used to determine the percentage of apoptotic and dead cells (% Cell death) in two different clones of shASS1 as compared to control. n = 3–4 biological replicates in each group as depicted in the graph. p value was calculated using Tukey multiple comparisons of means. p = 0.016 for control vs. shASS1#1; p = 0.028 for control vs. shASS1#2. f, Left panel- ASS1 is not required for proliferation of glucose-proficient lung cancer cells. A549 cells (expressing either ASS1-shRNA or control GFP-shRNA) were grown for 72 h. Survival was measured by crystal violet staining. Right panel- A549 cells with shASS1 and ASS1 OE were grown in nutrient deficient medium with and without MDLA. Survival was measured using quantified crystal violet staining. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.038. g, Bar plot showing the association between ASS1 expression and patient survival. Hazard ratio is shown in log10 scale (Y-axis) for 25 cancer types (those cancer types with statistical significance based on logrank test is marked black). The number of samples for each cancer type are listed in the Source Data.
Extended Data Fig. 2 ASS1 promotes nutrient-free cancer cell survival through pyruvate utilization for serine and glycine synthesis.
a, ASS1 is required for pyruvate utilization for gluconeogenesis in glucose-starved lung cancer cells. A549 cells (expressing either ASS1-shRNA or control GFP-shRNA) were grown in a glucose and serum-free medium with or without the addition of 1 mM pyruvate for 72 h. Survival was measured using crystal violet staining. n = 3 biological replicates in each group. p value was calculated using matched one-sided Wilcoxon ranksum test. P = 0.05 in control. b, Pyruvate accumulates in ASS1-deficient cancer cells under low-nutrient conditions. A549 cells (expressing either ASS1-shRNA or control GFP-shRNA) were grown in the presence or absence of glucose and serum for 24 h and quantified by liquid chromatography–mass spectrometry (LC-MS/MS) metabolomic profiling. n = 6 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.013 for the difference between the groups under no glucose state. c, Glycine is produced in nutrient-starved lung cancer cells when glucose is depleted. A549 cells were grown with or without glucose and serum (Low Nutrient) for 48 h. Cells were incubated with [U-13C] glutamine for the last 6 h of culture. Shown for glycine are the total levels quantified by GC/MS as the sum of areas under the curve (AUC) measurement for the different masses/ M + 0; shown for glucose are total levels as AUC. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 3.1E-05 for glycine labelling and 1.3E-04 for total glucose levels under low nutrient conditions compared to regular medium. d, ASS1 supports serine and glycine production from glutamine under nutrient deprivation. A549 cells were grown for 48 h followed by incubation with [U-13C] glutamine for 6 h. Shown is the isoropologue distribution of [U-13C]-labelled serine and glycine as quantified by GC/MS under low nutrients vs. normal medium in control vs. shASS1. n = 4 biological replicates in each group. p value was calculated using two-tailed student’s t-test. For serine: p = 0.0003 in M + 2 and 0.018 in M + 3; For glycine: p = 0.014 for M + 1. e, ASS1 inhibition does not alter total serine (left panel) or glycine levels (right panel). For the total levels of serine and glycine, A549 cells were grown for 48 h with or without glucose and serum followed by culture with [U-13C] glutamine for the last 6 h of incubation. Shown are total levels quantified in GC/MS by area under the curve (AUC) measurement. n = 4 biological replicates in each group. p value was calculated using two-tailed student’s t-test. f, ASS1 supports the synthesis of gluconeogenesis intermediates and glycine production under glucose deprivation.4T1 cells were grown with no glucose for 48 h. Metabolites were analysed by LC-MS. PEP was measured by AUC normalized to protein concentration. Shown is the increment in PEP levels in glucose-deprived cells relative to cells grown in normal conditions (Left panel), and the levels of [U-13C]-labelled mass pools/M + 0 of 3PG and glycine, as quantified by LC/MS (Right panel). n = 3–5 biological replicates in each group as depicted in the graphs. p value was calculated using two-tailed student’s t-test. For PEP p = 0.04; for 3PG p = 0.017 in no glucose; for glycine p = 0.005.
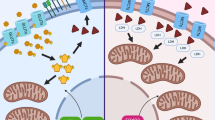
Extended Data Fig. 3 ASS1 promotes gluconeogenesis via nitrosylation.
a, NO donors (SNAP), rescue survival of starved ASS1-inhibited lung cancer cells. A549 cells were grown for 72 h with and without 10 mM SNAP. Survival of shASS1 cells relative to control cells was determined using crystal violet staining. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.047. b, NO is required for proliferation of ASS1-overexpressing cancer cells. 4T1 cells were grown for 48 h with or without the addition of 10mM L-NAME. Cell viability was measured using crystal violet staining. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.015. c, Quantification of the westrn shown in Fig. 3g demonstrating ASS1 necessity for starvation-induced S-nitrosylation of gluconeogenesis enzymes in nutrient-deficient lung cancer cells. A549 cells (expressing either ASS1-shRNA or control GFP-shRNA) were grown in the absence of glucose and serum for 48 h. Detection of protein nitrosylation was performed using the SNO-RAC method followed by a western blot. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.036 for PC and 0.002 for PCK2. d, S-nitrosylation of gluconeogenic enzymes in nutrient-proficient lung cancer cells. A549 cells were grown in the presence of glucose and serum for 48 h. Detection of protein nitrosylation was performed using the SNO-RAC method followed by a western blot and quantification. e, ASS1 is required for starvation-induced S-nitrosylation of gluconeogenic enzymes in livers of fasting mice. ASS1f/f Alb-CRE and control mice were fed either regularly or fasted for 16 h. Detection of protein nitrosylation in the liver was performed using the SNO-RAC method followed by a western blot and quantification. n = 3 mice in each group. p value was calculated using two-tailed student’s t-test. p = 0.042.
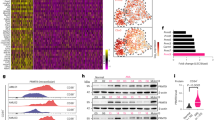
Extended Data Fig. 4 ASS1 expression in cancer is part of the transcriptional signature activated by c-Myc in response to nutrient deficiency.
ASS1 expression is significantly higher when c-MYC nutrient deprivation signature is high (top tertile, red) vs low (bottom tertile, blue) in TCGA breast, lung (LUAD, LUSC) and colon cancer. Wilcoxon ranksum P = 4E-41, 0.03, 0.3, and 0.12, respectively. * denotes P < 0.05, ****P < 0.0001. b, A western blot showing c-Myc levels in 4T1 cells grown with or without glucose for 24 h. c, DAF-FM measurement of NO levels– A549 cells were grown in glucose and serum free medium for 20hrs and treated with 10 μM 10074-G5 (c-Myc inhibitor, Sigma). Intracellular NO was detected by DAF-FM labeling and analyzed on flow cytometer. d, RT-PCR analysis showing elevation in the mRNA levels of PHGDH and PSPH in A549 cancer cells grown under no-glucose conditions in a c-Myc dependent manner. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. In PHGDH: p = 0.022 for control vs. no glucose and 0.007 for control to no glucose+10074-G5. In PSPH: p = 0.047 for control to no glucose and 0.021 for control to no glucose+10074-G5 (c-MYC inhibitor). e, A549 cells were grown in full medium (glucose +) or in glucose free medium (glucose –) and treated with 10 µM 10074-G5 (c-Myc inhibitor) for 48hrs. ASS1 and C-Myc levels were measured using western blotting. f, Gluconeogenic enzymes’ (PC + PCK1 + PCK2 + FBP1 + FBP2) expression is significantly higher in high ASL + ASS1 + NOS1 samples (top tertile) in breast, lung (LUAD + LUSC) and colon cancer compared to low ASL + ASS1 + NOS1 samples (bottom tertile). * denotes P < 0.05.
Extended Data Fig. 5 Restricting purine synthesis decreases the purine to pyrimidine ratio and inhibits growth of high ASS1 cancer cells.
a, Purine/pyrimidine (R/Y) ratio is increased upon nutrient deprivation in human colon cancer cells. MC38 cells were grown in either regular medium or in medium without glucose for 24 h. Nucleotide levels were measured by LC-MS. Purine/pyrimidine ratio was calculated by dividing (AMP + GMP) AUC by (UMP + CMP) AUC. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.0004. b, 4T1 cells with and without shASS1 were grown in glucose free medium either supplemented or not with 10mM L-Name for 48hrs. Nucleotide levels were measured by LC-MS. Purine/pyrimidine ratio was calculated by dividing the sum levels of (AMP + GMP) by the sum levels of (UMP + CMP + TMP). n = 2–3 biological replicates in each group as depicted in the graph. p value was calculated using two-tailed student’s t-test. p = 0.027 for control and 0.011 for shASS1. c, Purine synthesis inhibition results in decreased purine/pyrimidine ratio in human colon cancer cells. MC38 cells were grown either with or without 300μM mizoribine for 24 h. Nucleotide levels were measured by LC-MS. Purine/pyrimidine ratio was calculated as in A. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.002. d, Purine synthesis inhibition restricts ASS1-overexpressing cancer cell proliferation. 4T1 (left panel) and MC38 cells (right panel) were grown for 24 h with or without 300μM mizoribine. Cell viability was measured by crystal violet staining. n = 3 biological replicates in each group. p value was calculated using two-tailed student’s t-test. p = 0.009 for 4T1 and 0.0009 for MC38.
Extended Data Fig. 6 Identification of ASS1 and PD1 in NSCLC explants and autologous T-.
a, Immunohistochemistry staining of ASS1 expression is positive in the NSCLC PDXs of both patients. b, The cultured primary CD8+ T cells were mostly PD1 positive in both patients previously reported to be resistant to anti-PD1 treatment before or when biopsy and blood samples were taken. c, A schematic summary diagram. ASS1 expression in cancer benefits cancer cells’ survival under limiting glucose conditions via S-nitrosylation of gluconeogenic enzymes and by promoting purine synthesis. Treating tumors expressing ASS1 with purine synthesis inhibitors restricts tumor growth and promotes response to immune checkpoint inhibitors.
Supplementary information
Supplementary Tables
Supplementary Tables 1–5.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and gating.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and FACS gating.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots and FACS gating.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
FACS gating.
Rights and permissions
About this article
Cite this article
Keshet, R., Lee, J.S., Adler, L. et al. Targeting purine synthesis in ASS1-expressing tumors enhances the response to immune checkpoint inhibitors. Nat Cancer 1, 894–908 (2020). https://doi.org/10.1038/s43018-020-0106-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0106-7
This article is cited by
-
Serine synthesis and catabolism in starved lung cancer and primary bronchial epithelial cells
Cancer & Metabolism (2024)
-
Cytoophidia: a conserved yet promising mode of enzyme regulation in nucleotide metabolism
Molecular Biology Reports (2024)
-
PSMD2 contributes to the progression of esophageal squamous cell carcinoma by repressing autophagy
Cell & Bioscience (2023)
-
Mutant p53 sustains serine-glycine synthesis and essential amino acids intake promoting breast cancer growth
Nature Communications (2023)
-
The combined signatures of the tumour microenvironment and nucleotide metabolism-related genes provide a prognostic and therapeutic biomarker for gastric cancer
Scientific Reports (2023)