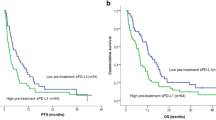
Abstract
Immune checkpoint blockade (ICB) provides clinical benefit to a subset of patients with cancer. However, existing biomarkers do not reliably predict treatment response across diverse cancer types. Limited data exist to show how serial circulating tumor DNA (ctDNA) testing may perform as a predictive biomarker in patients receiving ICB. We conducted a prospective phase II clinical trial to assess ctDNA in five distinct cohorts of patients with advanced solid tumors treated with pembrolizumab (NCT02644369). We applied bespoke ctDNA assays to 316 serial plasma samples obtained at baseline and every three cycles from 94 patients. Baseline ctDNA concentration correlated with progression-free survival, overall survival, clinical response and clinical benefit. This association became stronger when considering ctDNA kinetics during treatment. All 12 patients with ctDNA clearance during treatment were alive with median 25 months follow up. This study demonstrates the potential for broad clinical utility of ctDNA-based surveillance in patients treated with ICB.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Anonymized WES data of tumor and match normals are available with controlled access approval through the European Genome-Phenome Archive under accession number EGAS00001003280. Source data for Figs. 1–6 can be found in Source data Figs. 1–6 files, and Source data for Extended Data Fig. 2–8 can be found in Source data Extended Data Fig. 2–8 files. The supporting variant level data for all figures are available in Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
Code availability
Code necessary to reproduce the core analyses are available without restrictions at https://github.com/pughlab/inspire-ctdna.
References
Bedognetti, D. et al. Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J. Immunother. Cancer 7, 131 (2019).
Hause, R. J., Pritchard, C. C., Shendure, J. & Salipante, S. J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22, 1342–1350 (2016).
Jenkins, R. W., Thummalapalli, R., Carter, J., Canadas, I. & Barbie, D. A. Molecular and genomic determinants of response to immune checkpoint inhibition in cancer. Annu. Rev. Med. 69, 333–347 (2018).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 515, 568–571 (2014).
Gibney, G. T., Weiner, L. M. & Atkins, M. B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 17, e542–e551 (2016).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Wolchok, J. D. et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin. Cancer Res. 15, 7412–7420 (2009).
Ribas, A., Chmielowski, B. & Glaspy, J. A. Do we need a different set of response assessment criteria for tumor immunotherapy? Clin. Cancer Res. 15, 7116–7118 (2009).
Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 379, 1754–1765 (2018).
Cabel, L. et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat. Rev. Clin. Oncol. 15, 639–650 (2018).
Anagnostou, V. et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res. 79, 1214–1225 (2019).
Cabel, L. et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann. Oncol. 28, 1996–2001 (2017).
Giroux Leprieur, E. et al. Circulating tumor DNA evaluated by Next-Generation Sequencing is predictive of tumor response and prolonged clinical benefit with nivolumab in advanced non-small cell lung cancer. Oncoimmunology. 7, e1424675 (2018).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Gray, E. S. et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 6, 42008–42018 (2015).
Lee, J. H. et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann. Oncol. 28, 1130–1136 (2017).
Lee, J. H. et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol. 4, 717–721 (2018).
Raja, R. et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin. Cancer Res. 24, 6212–6222 (2018).
Moding, E. J. et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 1, 176–183 (2020).
Clouthier, D. L. et al. An interim report on the investigator-initiated phase 2 study of pembrolizumab immunological response evaluation (INSPIRE). J. Immunother. Cancer 7, 72 (2019).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Magbanua, M. J. M. et al. Circulating tumor DNA in neoadjuvant treated breast cancer reflects response and survival. Preprint at medRxiv https://doi.org/10.1101/2020.02.03.20019760 (2020).
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
Reinert, T. et al. Analysis of of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Lai, Z. et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108 (2016).
Salipante, S. J., Scroggins, S. M., Hampel, H. L., Turner, E. H. & Pritchard, C. C. Microsatellite instability detection by next generation sequencing. Clin. Chem. 60, 1192–1199 (2014).
McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra254 (2015).
Acknowledgements
Major funding support for the project was made possible by the Princess Margaret Cancer Foundation, Ontario Institute for Cancer Research and Terry Fox Research Institute. We thank Merck for contributing the study drug for the clinical trial. S.V.B. and T.J.P. were supported by the Gattuso-Slaight Personalized Cancer Medicine Fund. S.Y.C.Y. was supported in part by the University of Toronto Medical Biophysics OSOTF Excellence Award. M.A.J.I. was supported in part by a fellowship through the BMO Chair in Precision Cancer Genomics. L.L.S. holds the BMO Chair in Precision Cancer Genomics. T.J.P. holds the Canada Research Chair in Translational Genomics and is supported by a Senior Investigator Award from the Ontario Institute for Cancer Research. We are grateful to R. Salari, H.-T. Wu and S. Dashner for their technical support and insightful discussions of the project. We gratefully acknowledge the individuals from the Princess Margaret Tumour Immunotherapy Program (https://www.pm-tumorimmunotherapyprogram.ca/), including the immune monitoring (M. Butler, D. Gray, V. Sotov and D. Liu) and tissue correlatives (V. Speers, A. Giesler and S. Hakgor) teams. We thank the staff of the Princess Margaret Genomics Centre (https://www.pmgenomics.ca), Bioinformatics and High-Performance Computing Core, and PM-OICR Translational Genomics Laboratory (https://labs.oicr.on.ca/translational-genomics-laboratory) for their expertise in generating the sequencing data used in this study. Additional infrastructure support from the Canada Foundation for Innovation, Leaders Opportunity Fund (32383); Ontario Ministry of Research and Innovation, Ontario Research Fund Small Infrastructure Program and Ontario Institute for Cancer Research (https://genomics.oicr.on.ca).
Author information
Authors and Affiliations
Contributions
S.V.B., L.L.S. and T.J.P. conceived the study. S.V.B., P.S.O., L.L.S. and T.J.P. secured funding. S.Y.C.Y. performed data analysis. M.A.J.I. reviewed the clinical data and performed data analysis. A.R.H., P.L.B., S.L., A.S., A.A.R. and L.L.S. accrued patients and supervised biospecimen sample collection. T.J.P., P.S.O., D.L.C. and D.T. supervised biospecimen sample processing by S.Y.C.Y., S.C.L., Y.H., I.C., K.M. and J.E. W.X. and Z.L. performed statistical analysis. S.S., M.L., B.Z., P.B., H.S. and A.A. conducted the ctDNA assays and provided technical expertise and scientific feedback. S.V.B., S.Y.C.Y., L.L.S., W.X., Z.L., D.T. and T.J.P. wrote the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages.
Corresponding authors
Ethics declarations
Competing interests
S.V.B. provides consultation for Bristol-Myers Squibb, receives research support from Nektar Therapeutics and is a coinventor on a patent application relating to circulating tumor DNA mutation detection technology (PCT/US2014/02502, licensed to Roche Molecular Diagnostics) and on a patent application relating to ctDNA methylation analysis technology (PCT/CA2018/000203). L.L.S. provides consultation for Merck (compensated), Pfizer (compensated), Celgene (compensated), AstraZeneca (compensated), Morphosys (compensated), Roche (compensated), GeneSeeq (compensated), Loxo (compensated), Oncorus (compensated), Symphogen (compensated), Seattle Genetics (compensated), GlaxoSmithKline (compensated), Voronoi (compensated), Treadwell Therapeutics (compensated), Arvinas (compensated), Tessa (compensated), Navire (compensated); receives research support (institutional) from Novartis, Bristol-Myers Squibb, Pfizer, Boerhinger-Ingelheim, GlaxoSmithKline, Roche/Genentech, Karyopharm, AstraZeneca/Medimmune, Merck, Celgene, Astellas, Bayer, Abbvie, Amgen, Symphogen, Intensity Therapeutics, Mirati, Shattucks and Avid; and is a stockholder in Agios (spouse) and Treadwell Therapeutics (spouse). S.S., M.L., P.B., B.Z., H.S. and A.A. are employees of Natera, Inc. T.J.P. provides consultation for Merck, Chrysalis Biomedical Advisors and the Canadian Pension Plan Investment Board, and receives research support from Roche.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CONSORT diagram for the study.
The numbers of patients included in the study and reasons for exclusion are shown.
Extended Data Fig. 2 Factors affecting somatic mutation detection in tumor tissue and plasma cell-free DNA.
a, Somatic mutation burden per patient are grouped by INSPIRE cancer cohorts: Squamous cell cancer of head and neck (SCCHN, cohort A), triple negative breast cancer (TNBC, cohort B), high grade serous ovarian cancer (HGSOC, cohort C), malignant melanoma (MM, cohort D), and mixed solid tumors (MST, cohort E). The N (patients) for each group is shown in the top axis. Two-sided Kruskal-Wallis test p-value is shown. b, c, Comparison of 16 mutations vs 1 mutation from sequencing data for detecting ctDNA. To evaluate the impact of measuring 16 mutations on the likelihood of detecting ctDNA, we randomly selected one of the 16 mutations from each of the 94 baseline cell-free DNA samples and repeated this with 10,000 iterations. (b) An example of one of the 10,000 iteration shows that ctDNA levels (MTM/mL) are correlated (Spearman R = 0.87) among N = 94 baseline cell-free DNA samples. Two-sided Spearman correlation test p-value is shown. (c) Histogram of the detection likelihood (sensitivity for ctDNA) from 94 baseline cell-free DNA samples when using a randomly selected single mutation (N = 10,000 iterations). The mean sensitivity (red dashed line) is 84%. The sensitivity using all 16 mutations (black dashed line) is 98%. d, Influence of tumor tissue source used for bespoke assay design on detectability of ctDNA. The number of detected variants in baseline cell-free DNA samples are shown according to whether the bespoke assay design was conducted from FFPE vs fresh-frozen tumor exomes. Total number of samples in each group shown on the top axis. Two-sided Wilcoxon rank sum test p-value is shown. Box plots represent the median, upper and lower quartiles of each distribution with whiskers showing the limits of the distribution (1.5 times the interquartile range).
Extended Data Fig. 3 Rationale for calculation of absolute ctDNA levels from multiple mutation targets.
Analytical validation of the ctDNA quantification approach was conducted using Seraseq ctDNA Mutation Mix v2 (SeraCare, Milford, MA, USA) at various concentrations. For a given sample containing 16 target mutations, the mean tumor molecules per mL of plasma (MTM/mL) was calculated using two approaches (y-axis) and compared to known concentrations (x-axis). The blue data points represent the per-sample measurements of mean MTM/mL calculated over detected targets (slope=0.86), while the green data points represent the per-sample measurements of mean MTM/mL calculated over all 16 tested targets (slope=0.98). The red (X=Y) dashed line illustrates the ideal (expected) scenario in which the calculated experimental ctDNA levels are equal to the expected MTM/mL. The numbers of technical replicates at each concentration are shown (n = 2-18). In addition, all 10 negative control samples had no detected mutations (not shown). Pearson correlation coefficients (R2) are shown. This analytical comparison indicates that mean MTM/mL calculated over all tested targets provides a more accurate representation of the true amount of ctDNA, particularly at low levels of ctDNA.
Extended Data Fig. 4 ctDNA levels in the INSPIRE sub-cohorts and correlation with RECIST tumor measurements.
ctDNA levels (MTM/mL) at baseline a, and at cycle 3 b, as well as the fold-change in ctDNA levels from baseline to cycle 3 (∆ctDNAC3) c, are shown for each of the sub-cohorts: Squamous cell cancer of head and neck (SCCHN, cohort A), triple negative breast cancer (TNBC, cohort B), high grade serous ovarian cancer (HGSOC, cohort C), malignant melanoma (MM, cohort D), and mixed solid tumors (MST, cohort E). Box plots represent the median, upper and lower quartiles of each distribution with whiskers showing the limits of the distribution (1.5 times the interquartile range). The N (patients) for each group is shown in the top axis. Two-sided Kruskal-Wallis test p-values are shown for (a–c). d, Relationship between ctDNAB and RECIST 1.1 target lesion sum of tumor measurements (N = 94 patients). Scatter plot with Spearman correlation coefficient (R) and two-sided p-value are shown. The poor correlation in this diverse cohort of advanced cancers suggests that ctDNA may provide complementary information to RECIST tumor measurements. Of note, RECIST target lesion sum of tumor measurements does not incorporate measurements from non-target lesions.
Extended Data Fig. 5 ∆ctDNAC3 for the INSPIRE sub-cohorts.
Clinical response is indicated by line color. The inset table summarizes ∆ctDNAC3 values among patients with distinct clinical responses in each sub-cohort. Closed circles indicate ctDNA positive samples and open circles indicate ctDNA negative samples; SD, stable disease; CR, complete response; PR, partial response; PD, progressive disease; B, baseline; C3, cycle 3; ND, not detected (below detection threshold).
Extended Data Fig. 6 Relationship between ctDNA, biomarkers, and prognosis.
a, Relationship between ctDNA metrics (ctDNAB or ∆ctDNAC3) and other candidate biomarkers (PD-L1 or TMB). Scatter plots with Spearman correlation coefficients (R) and p-values are shown. Top row: N = 94 patients with both ctDNAB and TMB levels; N = 93 patients with both ctDNAB and PD-L1 levels. Bottom row: N = 72 patients with ∆ctDNAC3, TMB, and PD-L1 levels. b, Performance of multivariable Cox models incorporating ctDNA metrics (ctDNAB and/or ∆ctDNAC3). Multivariable Cox models for OS (left) or PFS (right) were generated. The baseline model incorporates cohort, PD-L1, and TMB. C-statistic is plotted for each model. Statistically significant differences (two-sided likelihood ratio test) are indicated.
Extended Data Fig. 7 Definitions of ctDNA dynamic profiles.
Serial ctDNA measurements in patients grouped according ctDNA dynamics (clearance at any time point, no clearance with all on-treatment values below baseline, no clearance with any on-treatment value above baseline). Triangles indicate values off the displayed range of the y-axis.
Extended Data Fig. 8 Landmark analyses of ctDNA clearance.
Kaplan-Meier curves of OS among patients classified as one of 3 groups (Clearance; Decrease from baseline; Increase from baseline) based on ctDNA at each indicated landmark (that is, C3, C6, C9, or C12).
Supplementary information
Supplementary Tables
Supplementary Tables 1–8.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
About this article
Cite this article
Bratman, S.V., Yang, S.Y.C., Iafolla, M.A.J. et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 1, 873–881 (2020). https://doi.org/10.1038/s43018-020-0096-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0096-5
This article is cited by
-
Nucleic acid-based drugs for patients with solid tumours
Nature Reviews Clinical Oncology (2024)
-
Tumor reactive γδ T cells contribute to a complete response to PD-1 blockade in a Merkel cell carcinoma patient
Nature Communications (2024)
-
Analytical evaluation of circulating tumor DNA sequencing assays
Scientific Reports (2024)
-
Multimodal detection of molecular residual disease in high-risk locally advanced squamous cell carcinoma of the head and neck
Cell Death & Differentiation (2024)
-
A multiparameter liquid biopsy approach allows to track melanoma dynamics and identify early treatment resistance
npj Precision Oncology (2024)