Abstract
Metabolic reprogramming is a core hallmark of cancer but it remains poorly defined in hepatocellular carcinogenesis (HCC). Here we show that hepatic aldolase B (Aldob) suppresses HCC by directly binding and inhibiting the rate-limiting enzyme in the pentose phosphate pathway, glucose-6-phosphate dehydrogenase (G6PD). A stage-dependent decrease of Aldob and increase of G6PD in human tumors are correlated with poor prognosis for patients with HCC. Global or liver-specific Aldob knockout promotes tumorigenesis in mice through enhancing G6PD activity and pentose phosphate pathway metabolism, whereas pharmacological inhibition or genetic knockdown of G6PD suppresses HCC. Consistently, restoration of Aldob in Aldob knockout mice attenuates tumorigenesis. We further demonstrate that Aldob potentiates p53-mediated inhibition of G6PD in an Aldob–G6PD–p53 complex. This scaffolding effect is independent of Aldob enzymatic activity. Together, our study reveals a new mode of metabolic reprogramming in HCC due to the loss of Aldob, suggesting a potential therapeutic strategy for HCC treatment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Microarray data of human HCC tissues were deposited at the Gene Expression Omnibus with accession number GSE74656. Source data for Figs. 1–6 and Extended Data Figs. 1–8 have been provided with the paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
Hatziapostolou, M. et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147, 1233–1247 (2011).
Park, E. J. et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140, 197–208 (2010).
Hay, N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer. 16, 635–649 (2016).
Levine, A. J. & Puzio-Kuter, A. M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344 (2010).
Li, B. et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature 513, 251–255 (2014).
Whittle, M. C. et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 161, 1345–1360 (2015).
Yeh, H. W. et al. PSPC1 mediates TGF-β1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat. Cell Biol. 20, 479–491 (2018).
DeBerardinis, R. J. & Thompson, C. B. Cellular metabolism and disease: what do metabolic outliers teach us?. Cell 148, 1132–1144 (2012).
DeBerardinis, R. J. & Chandel, N. S. Fundamentals of cancer metabolism. Sci. Adv. 2, e1600200 (2016).
Du, W. et al. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat. Cell Biol. 15, 991–1000 (2013).
Chen, L. et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat. Metab. 1, 404–415 (2019).
Lu, M. et al. Elevated G6PD expression contributes to migration and invasion of hepatocellular carcinoma cells by inducing epithelial-mesenchymal transition. Acta Bioch. Bioph. Sin. 50, 370–380 (2018).
Dore, M. P., Vidili, G., Marras, G., Assy, S. & Pes, G. M. Inverse association between glucose‒6‒phosphate dehydrogenase deficiency and hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 19, 1069–1073 (2018).
Hu, H. et al. Changes in glucose-6-phosphate dehydrogenase expression results in altered behavior of HBV-associated liver cancer cells. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G611–G622 (2014).
Mele, L. et al. A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 9, 572 (2018).
Lebherz, H. G. & Rutter, W. J. Distribution of fructose diphosphate aldolase variants in biological systems. Biochemistry 8, 109–121 (1969).
Rutter, W. J., Rajkumar, T., Penhoet, E., Kochman, M. & Valentine, R. Aldolase variants: structure and physiological significance. Ann. NY Acad. Sci. 151, 102–117 (1968).
Koo, H.-Y. et al. Dietary fructose induces a wide range of genes with distinct shift in carbohydrate and lipid metabolism in fed and fasted rat liver. Biochim. Biophys. Acta 1782, 341–348 (2008).
Bu, P. et al. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 27, 1249–1262 (2018).
Oppelt, S. A., Sennott, E. M. & Tolan, D. R. Aldolase-B knockout in mice phenocopies hereditary fructose intolerance in humans. Mol. Genet. Metab. 114, 445–450 (2015).
Chang, Y.-C., Yang, Y.-C., Tien, C.-P., Yang, C.-J. & Hsiao, M. Roles of aldolase family genes in human cancers and diseases. Trends Endocrin. Metab. 29, 549–559 (2018).
Penhoet, E., Rajkumar, T. & Rutter, W. J. Multiple forms of fructose diphosphate aldolase in mammalian tissues. Proc. Natl Acad. Sci. USA 56, 1275–1282 (1966).
Zhang, C. S. et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017).
Hai, H. et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016).
Bu, P. et al. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 27, 1249–1262 (2018).
Wang, Y. et al. Identification of four isoforms of aldolase B down-regulated in hepatocellular carcinoma tissues by means of two-dimensional western blotting. In Vivo 25, 881–886 (2011).
Peng, S. Y., Lai, P. L., Pan, H. W., Hsiao, L. P. & Hsu, H. C. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol. Rep. 19, 1045–1053 (2008).
Tao, Q. F. et al. Aldolase B inhibits metastasis through ten-eleven translocation 1 and serves as a prognostic biomarker in hepatocellular carcinoma. Mol. Cancer 14, 170 (2015).
Jiang, P., Du, W. & Yang, X. A critical role of glucose-6-phosphate dehydrogenase in TAp73-mediated cell proliferation. Cell Cycle 12, 3720–3726 (2013).
Ma, X. et al. Polo-like kinase 1 coordinates biosynthesis during cell cycle progression by directly activating pentose phosphate pathway. Nat. Commun. 8, 1506 (2017).
Zhang, C., Zhang, Z., Zhu, Y. & Qin, S. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med. Chem. 14, 280–289 (2014).
Lanaspa, M. A. et al. Ketohexokinase C blockade ameliorates fructose-induced metabolic dysfunction in fructose-sensitive mice. J. Clin. Invest. 128, 2226–2238 (2018).
Oppelt, S. A., Sennott, E. M. & Tolan, D. R. Aldolase-B knockout in mice phenocopies hereditary fructose intolerance in humans. Mol. Genet. Metab. 114, 445–450 (2015).
Jiang, P. et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316 (2011).
Poulain, L. et al. High mTORC1 activity drives glycolysis addiction and sensitivity to G6PD inhibition in acute myeloid leukemia cells. Leukemia 31, 2326–2335 (2017).
Au, S. W., Gover, S., Lam, V. M. & Adams, M. J. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure 8, 293–303 (2000).
Dalby, A. R., Tolan, D. R. & Littlechild, J. A. The structure of human liver fructose-1,6-bisphosphate aldolase. Acta Crystallogr. D Biol. Crystallogr. 57, 1526–1533 (2001).
Santamaria, R. et al. Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at arginine 303 in human liver aldolase. Biochem. J. 350, 823–828 (2000).
Pusapati, R. V. et al. mTORC1-dependent metabolic reprogramming underlies escape from glycolysis addiction in cancer cells. Cancer Cell 29, 548–562 (2016).
Lewis, C. A. et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014).
Fan, J. et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 (2014).
Ying, H. Q. et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012).
Boroughs, L. K. & DeBerardinis, R. J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351–359 (2015).
DelNero, P. et al. Cancer metabolism gets physical. Sci. Transl. Med. 10, eaaq1011 (2018).
Hitosugi, T. et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 22, 585–600 (2012).
Goncalves, M. D. et al. High-fructose corn syrup enhances intestinal tumor growth in mice. Science 363, 1345–1349 (2019).
Jiang, P., Du, W. & Wu, M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 5, 592–602 (2014).
Ma, L. et al. Control of nutrient stress-induced metabolic reprogramming by PKCζ in tumorigenesis. Cell 152, 599–611 (2013).
Nanchen, A., Fuhrer, T. & Sauer, U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 358, 177–197 (2007).
van Winden, W. A., Wittmann, C., Heinzle, E. & Heijnen, J. J. Correcting mass isotopomer distributions for naturally occurring isotopes. Biotechnol. Bioeng. 80, 477–479 (2002).
Portnoy, V. A. et al. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655. Appl. Environ. Microbiol. 76, 6529–6540 (2010).
Millard, P., Letisse, F., Sokol, S. & Portais, J. C. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296 (2012).
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Shen, X. et al. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 10, 1516 (2019).
Hu, L. et al. Association of vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene 23, 298–302 (2004).
Cai, X. et al. Background progenitor activation is associated with recurrence after hepatectomy of combined hepatocellular-cholangiocarcinoma. Hepatology 56, 1804–1816 (2012).
Acknowledgements
We acknowledge the help from D. Li at SIBS, CAS, A. D Richardson from Sanford Burnham Medical Research Institutes, S.-Cai Lin and S. Lin at Xiamen University and J. Rabinowitz at Princeton University. This work was financially supported by grants from Chinese National Natural Science Foundation: 31671231 and 91857112 to H.Y.Y.; 31401015 to Y.Z.T.; 61134013, 91029301, 81221061, 81125018 to S.Q.C. and Chinese Ministry of Science and Technology MOST (2016YFD0400205) to H.Y.Y. We thank the Mass Spectrometry platform, Molecular Biology/Biochemistry/cell Technology platform, Experimental Animal platform and Biological Sample Pathology Analysis platform at SINH, CAS.
Author information
Authors and Affiliations
Contributions
H.Y.Y. and Y.Z.T. designed this study. M.L., Y.Z.T., X.X.H., H.M.Y., R.S., Y.F.T., Y.J.Z., G.J.L., N.N.W., X.Q.N., Y.L., Z.M.H., C.Z.Y. and Q.C.T. performed experiments. Y.Z.T., H.Y.Y. and M.L. wrote the paper. W.X.G., B.Y.J., N.L., F.Z. and S.Q.C. obtained and provided clinical samples. H.Z., T.L., S.X.L. and J.Y. isolated primary tumor cells. X.S. performed untargeted metabolomics experiment. Z.Y. and Z.L. helped with metabolites measurement by LC–MS platform. S.C.Z., H.W.Z. and J.P.D. contributed to studies of protein–protein interaction.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
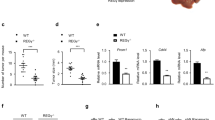
Extended Data Fig. 1 HCC patients with low Aldob and high G6PD expression exhibit aggressive tumor clinical pathologic features.
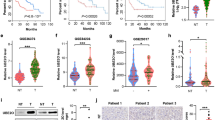
a, Hierarchical clustering of the gene expression patterns in matched normal liver, tumor and PVTT tissues from HCC patients (n = 5 HCC patients). Microarray analysis include 49,000 genes. b, Glycolysis and PPP pathways highlighting the roles of Aldob and G6PD. HK, hexokinase; PGI, phosphoglucose isomerase; PFKL, 6-phosphofructokinase, liver type; Aldob, fructose-1,6-bisphosphophate aldolase B; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F-1,6-BP, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; enzymes and metabolites in PPP: G6PD, glucose-6-phosphate dehydrogenase; PGD, phosphogluconate dehydrogenase; TKT, transketolase, 6PG, 6-phosphate gluconate; RU5P, ribulose 5-phosphate; GSH, glutathione; RX5P, xylulose 5-phosphate; E4P, erythrosine 4-phosphate; R5P, ribose 5-phosphate; S7P, sedoheptulose-7-phosphate. TCA cycles, tricarboxylic acid cycles. c, Representative Immunohistochemistry staining of Aldob and G6PD in matched tissues from HCC patients (n = 22, Scale bars, 25 □m). d, A reverse correlation of Aldob protein expression with G6PD (n = 166 HCC patients). e, Univariate analysis of hazard ratios (HR) of Aldob or G6PD protein expression (n = 184 HCC patients). f, Multivariant analysis of hazard ratios (HR) of a combination of Aldob with G6PD together including encapsulation, tumor size and macro-vascular invasion (n = 166 HCC patients). g-h, Representative TMA images with Aldoa (g) and Aldoc (h) antibody. i-j, Quantification of protein expression score of the matched normal tissues minus tumors from 184 HCC tissue microarray data with Aldoa (i) and Aldoc (j) antibody. Protein expression intensity is classified into negative (0-20 score), weak (21-100 score), moderate (101-180 score), strong (181-255 score). The calculation formula: (weak + moderate + strong)/(weak + moderate + strong + negative) x 100%. k-l, Kaplan-Meier survival curves in HCC patients with Aldoa (k, n = 214) and Aldoc (i, n = 203) high and low expression (patients are divided into two groups with high (>50) and low (< 50%) Aldoa or Aldoc expression). m, TMA antibody characterization of Aldob, G6PD, Aldoa, Aldoc (magnification: 200×). c, g, h, m, Experiments were repeated with three times independently with similar results; data of one representative experiment are shown. Statistical significance was determined by Cox Proportional Hazards Analyses (e, f), or log-rank test (k, l,).
Extended Data Fig. 2 Aldob expression inhibits glucose metabolism, tumor growth and metastasis.
a, Expression of Aldob decreases cell proliferation in Huh7 (n = 6 biologically independent samples). b, Knockdown of Aldob promotes cell proliferation in the primary liver cancer cell LIXC501, isolated from HCC patients (n = 6 biologically independent samples). c-d, Glucose consumption (c) and lactate production (d) with exogenous Aldob expression in Huh7 or knockdown of Aldob in LIXC501 cells (n = 3 biologically independent samples). e-g, Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) are used to measure metabolic changes with exogenous Aldob expression: Aldob expression significantly decreases oxygen consumption (e-f) and Glycolysis ratio (g) (n = 3 biologically independent samples). h-i, Cell migration in Aldob overexpression (h) and Aldob knockdown (i) cells (n = 4 biologically independent samples). j, Representative images and quantification of Ki67, β-catenin, and VEGFA expression (n = 7 mice per group). IHC score = intensity x area. The intensity is graded as 0, 1, 2, 3. The area of positive staining is assessed by 0, 1-25%, 26-50%, 51-75%, 76-100%, corresponding to 0, 1, 2, 3, 4 grades, respectively, scale bars, 50 μm. k, Representative images and incidence of lung metastasis, scale bars, 50 μm. H& E staining is performed to analyze lung metastasis (n = 5 male mice per group). a-d, f-k, Data represent the mean ± SD. Statistical significance was determined by two-way ANOVA (a, b), or two-tailed unpaired t-test (c, d, f-k). a, b, i, Experiments of immunoblots were repeated with four times independently with similar results; data of one representative experiment are shown. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 2.
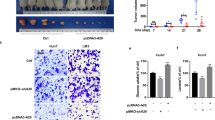
Extended Data Fig. 3 Global Aldob knockout (KO) promotes tumorigenesis in mice.
a, A 17 bp deletion (from 176 to192) of Aldob gene destroys Aldob protein expression using TALENs technique, and Western Blot of Aldob expression. b, WT and Aldob KO mice fed a fructose free diet do not show weight difference during the experimental period (n = 12 male mice per group). c, Aldolase enzymatic activity in WT and Aldob KO liver tissues (n = 3 biologically independent samples). d, Aldolase enzymatic activity in human HCC tissues (n = 3 biologically independent samples). e, Representative images of WT and Aldob KO livers and H&E histology of tumor nodules at 28th week (scale bars, 50 μm). f, Liver/body weight at 28th week (n = 8 male mice per group). g, Incidence of tumor with >5 mm in WT and Aldob KO mouse livers at 40th week (n = 12 male mice per group). h-i, Representative images (h) and IHC quantification of Aldob, Ki67, VEGFA and β-catenin expressions (i) (n = 10 mice per group, average of 3 IHC slices from each mouse used for statistical analysis) in mouse liver at 40th week (IHC score = intensity x area. The intensity was graded as 0, 1, 2, 3. The area of positive staining was assessed by 0, 1-25%, 26-50%, 51-75%, 76-100%, corresponding to 0, 1, 2, 3, 4 grades, respectively, scale bars, 50 μm). j, Differential metabolite analysis using untargeted metabolomics on WT and Aldob KO mice tumor at the age of 40th week by Triple TOF 6600 MS (n = 6 WT mice, n = 5 KO mice). k, Pathway enrichment analysis in metabolomics (n = 6 WT mice, n = 5 KO mice). l, Relative GSH/GSSG ratios using LC-MS (n = 7 male mice per group). b-d, f, i, l, Data represent the mean ± SD. Statistical significance was determined by two-way ANOVA (b), or two-tailed unpaired t-test (c, d, f, i, l). a, e, Experiments were repeated with three times independently with similar results; data of one representative experiment are shown. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 Restoration of Aldob expression in L-Aldob−/− mice attenuates tumor formation.
a, Genotyping and generation of L-Aldob−/− mice. b, Incidence of tumors >5 mm in Aldobf/f + CK AAV (n = 6 male mice), Aldobf/f + Aldob AAV (n = 5 male mice), L-Aldob-/- + CK AAV (n = 6 male mice) and L-Aldob-/- + Aldob AAV (n = 6 male mice) groups. c, Representative images of H&E staining (scale bars, 50 μm). d-e, Representative images of Ki67 staining and quantification in Aldobf/f + CK AAV (n = 6 male mice), Aldobf/f + Aldob AAV (n = 5 male mice), L-Aldob-/- + CK AAV (n = 6 male mice) and L-Aldob-/- + Aldob AAV (n = 6 male mice, average of 5 Ki673 IHC slices from each mouse used for statistical analysis) groups (scale bars, 50 μm). f-g, Relative NADPH/NADP+ ratios (f) and GSH/GSSG ratios (g) analyzed by targeted LC-MS/MS. Black: Aldobf/f + CK AAV (n = 6 male mice), Brown: Aldobf/f + Aldob AAV (n = 5 male mice), Red: L-Aldob-/-+CK AAV (n = 6 male mice), Blue: L-Aldob-/- + Aldob AAV (n = 6 male mice). h, Heatmap of the changes of metabolites in four groups of mouse liver tumor tissues by LC-MS, Aldobf/f + CK AAV (n = 4 male mice), Aldobf/f + Aldob AAV (n = 3 male mice), L-Aldob-/-+CK AAV (n = 4 male mice), L-Aldob-/- + Aldob AAV (n = 4 male mice). Heatmap is generated in MetaboAnalyst (https://www.metaboanalyst.ca) using metabolites abundance after log transformation. e-g, Data represent the mean ± SD. Statistical significance was determined by two-tailed unpaired t-test (e-g). c, d, Experiments were repeated with three times independently with similar results; data of one representative experiment are shown.
Extended Data Fig. 5 G6PD knockdown inhibits PPP metabolism in human liver cancer cells and tumor growth in vivo.
a-h, G6PD Knockdown inhibits G6PD activity (a) (n = 3 biologically independent samples), Cell proliferation (b) (n = 4 biologically independent samples), Cell migration (c) (n = 4 biologically independent samples), Oxidative PPP metabolism (d) (n = 3 biologically independent samples), Cellular NADPH production (e) (n = 3 biologically independent samples), NADPH/NADP+ ratio (f) (n = 3 biologically independent samples), TCA metabolism (g-h) (n = 3 biologically independent samples). i-j, The tumor images in xenograft mouse model injected with Huh7-shNT and Huh7-shG6PD cells (i) and tumor weights (j) (n = 6 male mice). k, Ki67 staining of tumor tissues and quantification (scale bars, 50 μm, n = 6 male mice, average of 5 Ki673 IHC slices from each mouse used for statistical analysis). a-h, j, k, Data represent the mean ± SD. Statistical significance was determined by two-tailed unpaired t-test (a, c-h, j, k), or two-way ANOVA (b). a-k, Experiments were repeated with three times independently with similar results; data of one representative experiment are shown. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 Pharmacological Inhibition of G6PD activity attenuates Aldob deficiency-induced tumorigenesis.
a, Experimental design of HCC mouse model using G6PD inhibitor (6AN + DHEA) on WT and global Aldob KO mice. b, Representative liver images of WT + solvent, WT + inhibitor, KO + solvent and KO + inhibitor. Black: WT + solvent mice, brown: WT + inhibitor, red: Aldob KO + solvent mice, purple: Aldob KO + inhibitor mice. c-e, Liver/body weight (c), Tumor numbers (d) and Max tumor volume (e) among four groups (n = 6 male mice per group). f, Representative images of Ki67 IHC staining and quantification (n = 6 mice per group, average of 4 Ki673 IHC slices from each mouse used for statistical analysis) in four groups (scale bars, 50 μm). g, Relative G6PD enzyme activity in mouse liver tissues (n = 6 male mice per group). h, Relative levels of PPP metabolites 6PG and R5P analyzed by targeted LC-MS analysis (n = 6 male mice per group). i, Relative ratios of NADPH/NADP + in mouse liver tissues (n = 6 male mice per group). j, Relative GSH/GSSG ratios in mouse liver tissues. GSH and NADPH analysis are performed on LC-MS platform (n = 6 male mice per group). c-j, Data represent the mean ± SD. Statistical significance was determined by two-tailed unpaired t-test (c-j).
Extended Data Fig. 7 Aldob inhibits G6PD activity and enhances p53-mediated G6PD inhibition.
a, Immunofluorescent staining shows co-localization of Aldob and G6PD in Huh7 Aldob cells (scale bars, 10 μm). b, Overview of Aldob and G6PD crystal structures and the potential interaction sites through charge-charge interactions. Dotted lines indicate an outstanding positively charged region on the Aldob surface and a negatively charged region on the G6PD surface. c, The enzymatic activity of purified recombinant Aldob and its mutants (n = 3 biologically independent samples). d, The effect of Aldob and its mutations on G6PD enzyme activity (n = 3 biologically independent samples). e, Western Blot analysis of Aldob expressions in stable Huh7-vector, Huh7-Aldob, Huh7-Aldob-mutant cell lines. f, Protein interaction measurement between G6PD and Aldoa or Aldoc. g, Protein expression and G6PD enzyme activity measurement after Aldoa overexpression (n = 5 biologically independent samples). h, GST pull-down of recombinant GST-p53, His-Aldob and His-G6PD. i, IP assays are performed in exogenous flag-tagged p53 and myc-tagged G6PD transfected into Huh7-vector, Huh7-Aldob and Huh7-Aldob-mutant cells. j, IP assays are performed using G6PD and p53 antibody in WT and p53 KD mice (p53 knockdown mice). c, d, g, Data represent the mean ± SD. Statistical significance was determined by two-tailed unpaired t-test (c, d, g). a, c-k, Experiments were repeated with five times independently with similar results; data of one representative experiment are shown. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Aldob inhibits PPP metabolism.
a, IP experiments are performed to detect the interactions between Aldob and p53 after transfection of flag Aldob-WT, flag Aldob-R149A, flag Aldob-R304A and flag Aldob-mutant plasmids. These cells are also used in b-f. b, Labelled NADPH fraction from 100% [3-2H] glucose (2 g/L [3-2H] glucose) using a TripleTOF 6600 MS platform (n = 3 biologically independent samples). c-f, Schematic diagram and labelled PPP metabolites G6P, 6PG, R5P from 100% [U-13C6] glucose (2 g/L [U-13C6] glucose) using a TripleTOF 6600 platform (n = 3 biologically independent samples). b, d-f, Data represent the mean ± SEM. Statistical significance was determined by two-tailed unpaired t-test (b, d-f). a-b, d-f, Experiments were repeated with three times independently with similar results; data of one representative experiment are shown. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 8.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1.
Statistical source data.
Source Data Fig. 2.
Statistical source data.
Source Data Fig. 2.
Unprocessed western blots and/or gels.
Source Data Fig. 3.
Statistical source data.
Source Data Fig. 3.
Unprocessed western blots and/or gels.
Source Data Fig. 4.
Statistical source data.
Source Data Fig. 4.
Unprocessed western blots and/or gels.
Source Data Fig. 5.
Statistical source data.
Source Data Fig. 5.
Unprocessed western blots and/or gels.
Source Data Fig. 6.
Statistical source data.
Source Data Extended Data Fig. 1.
Statistical source data.
Source Data Extended Data Fig. 2.
Statistical source data.
Source Data Extended Data Fig. 2.
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3.
Statistical source data.
Source Data Extended Data Fig. 3.
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4.
Statistical source data.
Source Data Extended Data Fig. 5.
Statistical source data.
Source Data Extended Data Fig. 5.
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6.
Statistical source data.
Source Data Extended Data Fig. 7.
Statistical source data.
Source Data Extended Data Fig. 7.
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 8.
Statistical source data.
Source Data Extended Data Fig. 8.
Unprocessed western blots and/or gels.
Rights and permissions
About this article
Cite this article
Li, M., He, X., Guo, W. et al. Aldolase B suppresses hepatocellular carcinogenesis by inhibiting G6PD and pentose phosphate pathways. Nat Cancer 1, 735–747 (2020). https://doi.org/10.1038/s43018-020-0086-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0086-7
This article is cited by
-
G6PD maintains the VSMC synthetic phenotype and accelerates vascular neointimal hyperplasia by inhibiting the VDAC1–Bax-mediated mitochondrial apoptosis pathway
Cellular & Molecular Biology Letters (2024)
-
G6PD and machine learning algorithms as prognostic and diagnostic indicators of liver hepatocellular carcinoma
BMC Cancer (2024)
-
Development of a promising PPAR signaling pathway-related prognostic prediction model for hepatocellular carcinoma
Scientific Reports (2024)
-
Lysyl hydroxylase LH1 promotes confined migration and metastasis of cancer cells by stabilizing Septin2 to enhance actin network
Molecular Cancer (2023)
-
A mitotic NADPH upsurge promotes chromosome segregation and tumour progression in aneuploid cancer cells
Nature Metabolism (2023)