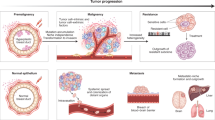
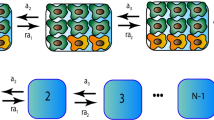
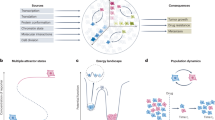
Uncovering and quantifying the laws of the evolutionary dynamics of cancer, in particular in the context of specific genetic lesions and in individual patients, has the potential to revolutionize precision oncology. Recent technological advances in the study of human cancer have increased access to in vivo human data and have thereby facilitated the confirmation or refutation of existing theoretical models. In this Perspective, we discuss recent work at the intersection of quantitative mathematical models of cancer evolution and patient data that provides insights into different stages of tumor evolution, including premalignant and malignant progression and response to therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bozic, I. & Nowak, M. A. Cancer. Unwanted evolution. Science 342, 938–939 (2013).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Tomasetti, C., Li, L. & Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017).
Podlaha, O., Riester, M., De, S. & Michor, F. Evolution of the cancer genome. Trends Genet. 28, 155–163 (2012).
Aktipis, C. A. & Nesse, R. M. Evolutionary foundations for cancer biology. Evol. Appl. 6, 144–159 (2013).
Ewens, W. J. Mathematical Population Genetics 1: Theoretical Introduction (Springer-Verlag, New York, 2004).
Desai, M. M., Fisher, D. S. & Murray, A. W. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394 (2007).
Good, B. H., Rouzine, I. M., Balick, D. J., Hallatschek, O. & Desai, M. M. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl Acad. Sci. USA 109, 4950–4955 (2012).
Neher, R. A., Russell, C. A. & Shraiman, B. I. Predicting evolution from the shape of genealogical trees. eLife 3, e03568 (2014).
Korolev, K. S. et al. Selective sweeps in growing microbial colonies. Phys. Biol. 9, 026008 (2012).
Waclaw, B. et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264 (2015).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168, 613–628 (2017).
Bozic, I., Gerold, J. M. & Nowak, M. A. Quantifying clonal and subclonal mutations during tumor evolution. PLOS Comput. Biol. 12, e1004731 (2016).
Fane, M. & Weeraratna, A. T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106 (2020).
de Visser, K. E. & Coussens, L. M. The inflammatory tumor microenvironment and its impact on cancer development. Contrib. Microbiol. 13, 118–137 (2006).
Taniguchi, K. & Karin, M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 18, 309–324 (2018).
Binnewies, M. et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018).
Fridman, W. H., Zitvogel, L., Sautès-Fridman, C. & Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 (2017).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Hu, Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019).
Khan, K. H. et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C Phase II colorectal cancer clinical trial. Cancer Discov. 8, 1270–1285 (2018).
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Salk, J. J., Schmitt, M. W. & Loeb, L. A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 19, 269–285 (2018).
Salk, J. J. et al. Ultra-sensitive TP53 sequencing for cancer detection reveals progressive clonal selection in normal tissue over a century of human lifespan. Cell Rep. 28, 132–144.e3 (2019).
Pekin, D. et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11, 2156–2166 (2011).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Silverbush, D. et al. Simultaneous integration of multi-omics data improves the identification of cancer driver modules. Cell Syst. 8, 456–466.e5 (2019).
Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
Friberg, S. & Mattson, S. On the growth rates of human malignant tumors: implications for medical decision making. J. Surg. Oncol. 65, 284–297 (1997).
Heuser, L., Spratt, J. S. & Polk, H. C. Jr. Growth rates of primary breast cancers. Cancer 43, 1888–1894 (1979).
Norton, L. A Gompertzian model of human breast cancer growth. Cancer Res. 48, 7067–7071 (1988).
Spratt, J. A., von Fournier, D., Spratt, J. S. & Weber, E. E. Decelerating growth and human breast cancer. Cancer 71, 2013–2019 (1993).
Mayneord, W. V. On a law of growth of Jensen’s rat sarcoma. Am. J. Cancer 16, 841–846 (1932).
Schrek, R. A quantitative study of the growth of the Walker rat tumor and the Flexner-Jobling rat carcinoma. Am. J. Cancer 24, 807–822 (1935).
Mandonnet, E. et al. Continuous growth of mean tumor diameter in a subset of grade II gliomas. Ann. Neurol. 53, 524–528 (2003).
Rodriguez-Brenes, I. A., Komarova, N. L. & Wodarz, D. Tumor growth dynamics: insights into evolutionary processes. Trends Ecol. Evol. 28, 597–604 (2013).
Gerlee, P. The model muddle: in search of tumor growth laws. Cancer Res. 73, 2407–2411 (2013).
Alexander, S., Koehl, G. E., Hirschberg, M., Geissler, E. K. & Friedl, P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem. Cell Biol. 130, 1147–1154 (2008).
Gimbrone, M. A. Jr., Leapman, S. B., Cotran, R. S. & Folkman, J. Tumor dormancy in vivo by prevention of neovascularization. J. Exp. Med. 136, 261–276 (1972).
Folkman, J. What is the evidence that tumors are angiogenesis dependent? J. Natl. Cancer Inst. 82, 4–6 (1990).
Naumov, G. N., Folkman, J. & Straume, O. Tumor dormancy due to failure of angiogenesis: role of the microenvironment. Clin. Exp. Metastasis 26, 51–60 (2009).
Demicheli, R. et al. An exponential-Gompertzian description of LoVo cell tumor growth from in vivo and in vitro data. Cancer Res. 49, 6543–6546 (1989).
Hart, D., Shochat, E. & Agur, Z. The growth law of primary breast cancer as inferred from mammography screening trials data. Br. J. Cancer 78, 382–387 (1998).
Talkington, A. & Durrett, R. Estimating tumor growth rates in vivo. Bull. Math. Biol. 77, 1934–1954 (2015).
Gruber, M. et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nature 570, 474–479 (2019).
Schmid, H. P., McNeal, J. E. & Stamey, T. A. Observations on the doubling time of prostate cancer. The use of serial prostate-specific antigen in patients with untreated disease as a measure of increasing cancer volume. Cancer 71, 2031–2040 (1993).
Armitage, P. & Doll, R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br. J. Cancer 8, 1–12 (1954).
Knudson, A. G. Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA 68, 820–823 (1971).
MacPherson, D. & Dyer, M. A. Retinoblastoma: from the two-hit hypothesis to targeted chemotherapy. Cancer Res. 67, 7547–7550 (2007).
Moolgavkar, S. H. & Knudson, A. G. Jr. Mutation and cancer: a model for human carcinogenesis. J. Natl Cancer Inst. 66, 1037–1052 (1981).
Luebeck, E. G. & Moolgavkar, S. H. Multistage carcinogenesis and the incidence of colorectal cancer. Proc. Natl Acad. Sci. USA 99, 15095–15100 (2002).
Meza, R., Jeon, J., Moolgavkar, S. H. & Luebeck, E. G. Age-specific incidence of cancer: phases, transitions, and biological implications. Proc. Natl Acad. Sci. USA 105, 16284–16289 (2008).
Beerenwinkel, N. et al. Genetic progression and the waiting time to cancer. PLOS Comput. Biol. 3, e225 (2007).
Bozic, I. et al. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl Acad. Sci. USA 107, 18545–18550 (2010).
Martens, E. A., Kostadinov, R., Maley, C. C. & Hallatschek, O. Spatial structure increases the waiting time for cancer. New J. Phys. 13, 115014 (2011).
Durrett, R., Foo, J., Leder, K., Mayberry, J. & Michor, F. Intratumor heterogeneity in evolutionary models of tumor progression. Genetics 188, 461–477 (2011).
Chowell, D. et al. Modeling the subclonal evolution of cancer cell populations. Cancer Res. 78, 830–839 (2018).
Tomasetti, C., Vogelstein, B. & Parmigiani, G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl Acad. Sci. USA 110, 1999–2004 (2013).
McFarland, C. D., Korolev, K. S., Kryukov, G. V., Sunyaev, S. R. & Mirny, L. A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl Acad. Sci. USA 110, 2910–2915 (2013).
Axelrod, D. & Kimmel, M. Branching Processes in Biology. (Springer, New York, 2015).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Petljak, M. et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020).
Baker, A. M. et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 8, 940–947 (2014).
Nicholson, A. M. et al. Fixation and spread of somatic mutations in adult human colonic epithelium. Cell Stem Cell 22, 909–918.e8 (2018).
Clevers, H. The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 (2011).
Vermeulen, L. et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995–998 (2013).
Klein, A. M., Brash, D. E., Jones, P. H. & Simons, B. D. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc. Natl Acad. Sci. USA 107, 270–275 (2010).
Maley, C. C. et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 64, 3414–3427 (2004).
Martinez, P. et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat. Commun. 7, 12158 (2016).
Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018).
Desai, P. et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018).
Abelson, S. et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Turajlic, S. et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 173, 595–610.e11 (2018).
Mitchell, T. J. et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 173, 611–623.e17 (2018).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Ling, S. et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl Acad. Sci. USA 112, E6496–E6505 (2015).
Tarabichi, M. et al. Neutral tumor evolution? Nat. Genet. 50, 1630–1633 (2018).
McDonald, T. O., Chakrabarti, S. & Michor, F. Currently available bulk sequencing data do not necessarily support a model of neutral tumor evolution. Nat. Genet. 50, 1620–1623 (2018).
Bozic, I., Paterson, C. & Waclaw, B. On measuring selection in cancer from subclonal mutation frequencies. PLOS Comput. Biol. 15, e1007368 (2019).
Sun, R. et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nat. Genet. 49, 1015–1024 (2017).
Williams, M. J. et al. Measuring the distribution of fitness effects in somatic evolution by combining clonal dynamics with dN/dS ratios. eLife 9, e48714 (2020).
Williams, M. J. et al. Quantification of subclonal selection in cancer from bulk sequencing data. Nat. Genet. 50, 895–903 (2018).
Marusyk, A. et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014).
Archetti, M., Ferraro, D. A. & Christofori, G. Heterogeneity for IGF-II production maintained by public goods dynamics in neuroendocrine pancreatic cancer. Proc. Natl Acad. Sci. USA 112, 1833–1838 (2015).
Gatenby, R. A. & Gillies, R. J. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer 8, 56–61 (2008).
Anderson, A. R. A., Weaver, A. M., Cummings, P. T. & Quaranta, V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127, 905–915 (2006).
Michor, F. et al. Dynamics of chronic myeloid leukaemia. Nature 435, 1267–1270 (2005).
Tang, M. et al. Dynamics of chronic myeloid leukemia response to long-term targeted therapy reveal treatment effects on leukemic stem cells. Blood 118, 1622–1631 (2011).
Wodarz, D. et al. Kinetics of CLL cells in tissues and blood during therapy with the BTK inhibitor ibrutinib. Blood 123, 4132–4135 (2014).
Schuh, A. et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 120, 4191–4196 (2012).
Burger, J. A. et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat. Commun. 7, 11589 (2016).
Ahn, I. E. et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood 129, 1469–1479 (2017).
Bozic, I. & Nowak, M. A. Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers. Proc. Natl Acad. Sci. USA 111, 15964–15968 (2014).
Landau, D. A. et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat. Commun. 8, 2185 (2017).
Körber, V. et al. Evolutionary trajectories of IDHWT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell 35, 692–704.e12 (2019).
Diaz, L. A. Jr. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Parseghian, C. M. et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann. Oncol. 30, 243–249 (2019).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Zhang, J., Cunningham, J. J., Brown, J. S. & Gatenby, R. A. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat. Commun. 8, 1816 (2017).
Kaznatcheev, A., Peacock, J., Basanta, D., Marusyk, A. & Scott, J. G. Fibroblasts and alectinib switch the evolutionary games played by non-small cell lung cancer. Nat. Ecol. Evol. 3, 450–456 (2019).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Schwarz, R. F. et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 12, e1001789 (2015).
Gao, R. et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 48, 1119–1130 (2016).
Acknowledgements
This work was supported in part by the National Cancer Institute (1R01CA155010-01A1, P01CA206978, U10CA180861). C.J.W. is a Scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Contributions
I.B. and C.J.W. conceived of and wrote this Perspective.
Corresponding author
Ethics declarations
Competing interests
C.J.W. is founder of Neon Therapeutics and is a member of its scientific advisory board and receives research funding from Pharmacyclics.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bozic, I., Wu, C.J. Delineating the evolutionary dynamics of cancer from theory to reality. Nat Cancer 1, 580–588 (2020). https://doi.org/10.1038/s43018-020-0079-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0079-6
This article is cited by
-
Tumor phylogeography reveals block-shaped spatial heterogeneity and the mode of evolution in Hepatocellular Carcinoma
Nature Communications (2024)
-
Neuroblastoma arises in early fetal development and its evolutionary duration predicts outcome
Nature Genetics (2023)
-
Autoencoder neural networks enable low dimensional structure analyses of microbial growth dynamics
Nature Communications (2023)
-
The coming decade in precision oncology: six riddles
Nature Reviews Cancer (2023)
-
Immunobiology of cancer stem cells and their immunoevasion mechanisms
Molecular Biology Reports (2023)