Abstract
Polyclonal metastases frequently arise from clusters of circulating tumor cells (CTCs). CTC clusters metastasize better than single CTCs, but the underlying molecular mechanisms are poorly understood. Here, we show that polyclonal metastatic seeds exhibit higher resistance to natural killer (NK) cell killing. Using breast cancer models, we observed higher proportions of polyclonal lung metastasis in immunocompetent mice compared with mice lacking NK cells. Depleting NK cells selectively increased monoclonal but not polyclonal metastases, suggesting that CTC clusters are less sensitive to NK-mediated suppression. Transcriptional analyses revealed that clusters have elevated expression of cell–cell adhesion and epithelial genes, which is associated with decreased expression of NK cell activating ligands. Furthermore, perturbing tumor cell epithelial status altered NK ligand expression and sensitivity to NK-mediated killing. Collectively, our findings show that NK cells can determine the fate of CTCs of different epithelial and mesenchymal states, and impact metastatic clonal evolution by favoring polyclonal seeding.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others

Data availability
The RNA-Seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession the code GSE144713. Previously published RNA-Seq data that were re-analyzed here are available under accession codes GSE51827, GSE111065, GSE104765 and GSE81955. Source data for Figs. 1–4 and 6 and Extended Data Figs. 1–5 and 7–9 are provided with the paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The custom code used to estimate the percentage local mixing of primary tumors was kindly provided by A. Ewald (Johns Hopkins University) and K. Cheung (Fred Hutchinson Cancer Research Center)7. Please direct any code requests to the original authors.
References
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis. Cell 168, 670–691 (2016).
Massagué, J. & Obenauf, A. C. Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Siegel, M. B. et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Invest. 128, 1371–1383 (2018).
Thiery, J. P., Acloque, H., Huang, R. Y. J. & Nieto, M. A. Epithelial–mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Dongre, A. & Weinberg, R. A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019).
Cheung, K. J. et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl Acad. Sci. USA 113, E854–E863 (2016).
Aiello, N. M. et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695.e4 (2018).
Reichert, M. et al. Regulation of epithelial plasticity determines metastatic organotropism in pancreatic cancer. Dev. Cell 45, 696–711.e8 (2018).
Klameth, L. et al. Small cell lung cancer: model of circulating tumor cell tumorospheres in chemoresistance. Sci. Rep. 7, 5337 (2017).
Krol, I. et al. Detection of circulating tumour cell clusters in human glioblastoma. Br. J. Cancer 119, 487–491 (2018).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Giuliano, M. et al. Perspective on circulating tumor cell clusters: why it takes a village to metastasize. Cancer Res. 78, 845–853 (2018).
Liu, X. et al. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov. 9, 96–113 (2019).
Gkountela, S. et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112.e14 (2019).
Jansson, S., Bendahl, P. O., Larsson, A. M., Aaltonen, K. E. & Rydén, L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer 16, 433 (2016).
Szczerba, B. M. et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 566, 553–557 (2019).
Kitamura, T., Qian, B. Z. & Pollard, J. W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 15, 73–86 (2015).
Meng, S. et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 10, 8152–8162 (2004).
Lanier, L. L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502 (2008).
Navarro, A. G., Björklund, A. T. & Chekenya, M. Therapeutic potential and challenges of natural killer cells in treatment of solid tumors. Front. Immunol. 6, 202 (2015).
López-Soto, A., Gonzalez, S., Smyth, M. J. & Galluzzi, L. Review control of metastasis by NK cells. Cancer Cell 32, 135–154 (2017).
Cheung, K. J., Gabrielson, E., Werb, Z. & Ewald, A. J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651 (2013).
Pearson, T. et al. Non-obese diabetic–recombination activating gene-1 (NOD–Rag1 null) interleukin (IL)-2 receptor common gamma chain (IL2rγ null) null mice: a radioresistant model for human lymphohaematopoietic engraftment. Clin. Exp. Immunol. 154, 270–284 (2008).
Cerwenka, A. & Lanier, L. L. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 16, 112–123 (2016).
Kasai, M. et al. In vivo effect of anti-asialo GM1 antibody on natural killer activity. Nature 127, 34–38 (1981).
Chen, Y. H., Chiu, N. M., Mandal, M., Wang, N. & Wang, C. R. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 6, 459–467 (1997).
Smyth, M. J., Crowe, N. Y. & Godfrey, D. I. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13, 459–463 (2001).
Vivier, E., Tomasello, E., Baratin, M., Walzer, T. & Ugolini, S. Functions of natural killer cells. Nat. Immunol. 9, 503–510 (2008).
Chockley, P. J. et al. Epithelial–mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer. J. Clin. Invest. 128, 1384–1396 (2018).
Lopez-Soto, A. et al. Epithelial–mesenchymal transition induces an antitumor immune response mediated by NKG2D receptor. J. Immunol. 190, 4408–4419 (2013).
Burk, U. et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 (2008).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008).
Korpal, M., Lee, E. S., Hu, G. & Kang, Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008).
Knezevic, J. et al. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene 34, 5997–6006 (2015).
Vance, R. E., Kraft, J. R., Altman, J. D., Jensen, P. E. & Raulet, D. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical MHC class I molecule Qa-1b. J. Exp. Med. 188, 1841–1848 (1998).
Carayannopoulos, L. N., Naidenko, O. V., Fremont, D. H. & Yokoyama, W. M. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169, 4079–4083 (2002).
Diefenbach, A., Hsia, J. K., Hsiung, M. Y. B. & Raulet, D. H. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur. J. Immunol. 33, 381–391 (2003).
Popa, N., Garmy, N. & Che, E. The NKG2D ligands RAE-1δ and RAE-1ε differ with respect to their receptor affinity, expression profiles and transcriptional regulation. PLoS ONE 5, e13466 (2010).
Chaudhury, A. et al. CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT. Nat. Commun. 7, 13362 (2016).
Kaiser, B. K. et al. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 174, 2878–2884 (2005).
Eisele, G. et al. TGF-β and metalloproteinases differentially suppress NKG2D ligand surface expression on malignant glioma cells. Brain 129, 2416–2425 (2006).
Taube, J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl Acad. Sci. USA 107, 15449–15454 (2010).
Labelle, M., Begum, S. & Hynes, R. O. Direct signaling between platelets and cancer cells induces an epithelial–mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590 (2011).
Ao, Z. et al. Identification of cancer-associated fibroblasts in circulating blood from patients with metastatic breast cancer. Cancer Res. 75, 4681–4687 (2015).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014).
Pastushenko, I. et al. Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018).
Li, W. & Kang, Y. Probing the fifty shades of EMT in metastasis. Trends Cancer 2, 65–67 (2016).
Tsai, J. H., Donaher, J. L., Murphy, D. A., Chau, S. & Yang, J. Spatiotemporal regulation of epithelial–mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Balsa-Canto, E., Henriques, D., Gábor, A. & Banga, J. R. AMIGO2, a toolbox for dynamic modeling, optimization and control in systems biology. Bioinformatics 32, 3357–3359 (2016).
Acknowledgements
We thank L. Metelitsa for providing the CD1d-KO mice and A. Ewald and K. Cheung for sharing ImageJ code for tumor color-mixing measurements. We also thank all members of the Zhang laboratory for discussion and support while developing this work. X.H.-F.Z. is supported by the Breast Cancer Research Foundation, NCI (CA227904), US Department of Defense (DAMD W81XWH-16-1-0073 and W81XWH-18-1-0574) and McNair Medical Institute. J.M.R. is supported by the NCI (CA148761) and CPRIT (RP170172). J.R.N. is supported by the NCI (CA190467) and The Samuel Waxman Cancer Research Foundation. A.S.K. is supported by P01 AI116501, R01 AI145108-01, IO1 IBX0104588A and R41 CA224520-01A1. The flow cytometry experiments were performed at the Cytometry and Cell Sorting Core at Baylor College of Medicine, with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672) and NIH (P30 CA125123 and S10 RR024574) and expert assistance from J. M. Sederstrom. Mosaic scanning was performed at the RNA In Situ Hybridization Core at Baylor College of Medicine, which is, in part, supported by a Shared Instrumentation grant from the NIH (1S10OD016167).
Author information
Authors and Affiliations
Contributions
H.C.L. and X.H.-F.Z. developed the concepts, designed the experiments and wrote the manuscript. H.C.L. performed the experiments. Z.X. performed the laser-capture microdissection and animal experiments. I.S.K. provided the cell lines and RNA-Seq data. B.P. assisted with the NK cell cytotoxicity assays. S.K. performed the fluorescence-activated cell sorting experiments. S.A., W.Z., A.M.M. and J.L. assisted with the animal experiments. S.M.H., A.S.K., J.R.N., S.P. and J.M.R. contributed to conceptual development and manuscript editing. H.W. analyzed the RNA-Seq data and discussed the project direction. X.H.-F.Z. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
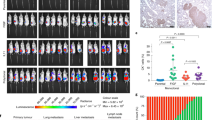
Extended Data Fig. 1 Mouse genetic background and immune deficiency affects the relative frequency of polyclonal metastases.
a, Representative microscopy image of AT3 and 4T1 primary tumor arising from equal numbers of GFP and mCherry-labelled cells. (Image representative of 5 mice) Scale bar, 1mm b, Representative microscopy images showing mixed-color AT3 lung lesions and immunohistochemistry staining for immune cell markers CD45, CD68 and CD11c. Yellow arrowheads indicate immune cells. (Image representative of 3 mice) Scale bar, 50 µm. c, Primary tumor weights of AT3 (B6, n = 12 mice; nude, n = 15 mice; NRG, n = 14 mice) and 4T1 (n = 15 mice per group) mammary gland tumors at the time of tumor removal. d, Quantification of local mixing % of red and green cells in AT3 mixed-colored primary tumors (n = 5 mice). e, Total lung metastatic lesion numbers for AT3 (B6, n = 12 mice; nude, n = 15 mice; NRG, n = 14 mice) and 4T1 tumors (n = 15 mice per group). f, Poisson regression analysis of mixed-colored tumor metastasis experiment for AT3 (B6, n = 12 mice; nude, n = 15 mice; NRG, n = 14 mice) and 4T1 tumors (n = 15 mice per group). g, Representative microscopy images and relative proportion (shown as a %) of single or mixed-colored lung metastasis lesions in NRG mice bearing LM2 tumors. (n = 7 mice) Scale bar, 50 µm. Mean ± s.d. shown. P values were calculated by two-tailed unpaired Student’s t-test (c,d), or two-tailed unpaired Mann–Whitney U-test (e).
Extended Data Fig. 2 Disseminated tumor cell clusters have higher metastatic advantage in wildtype and nude mice compared to NRG mice.
a, Schematic diagram for preparation of single cell and clusters for tail vein injection. b, Representative microscope images of single cells and clusters from GFP-labelled AT3 and 4T1 cells (Representative of 5 images/ group). Scale bar, 100 µm. c, d, Lung metastasis growth curve and representative whole body bioluminescence images of tail vein-injected c) AT3 cells (n = 5 mice per group) and d) 4T1 cells (n = 5 mice per group, except Balb/c single cells, n = 4) into NRG, nude and C57BL/6 or BALB/c mice. NRG mice bearing AT3 mice died before the other mouse strains. Mean ± s.e.m. shown. P values were calculated by two-way repeated measures ANOVA of log2 transformed growth curves (c, d).
Extended Data Fig. 3 NK cell depletion selectively increases lung metastasis growth by monoclonal seeding.
a, Treatment schematic and weights of AT3 primary tumors in Ctrl, anti-CSF-1R or anti-asialo GM1-treated mice, at the time of tumor removal (n = 15 mice per group). b, Representative flow plots of (top) blood NK cells from Ctrl and anti-ASGM1 antibody treated mice and (bottom) blood monocytes from Ctrl and anti-CSF1R antibody treated mice (n = 3 mice). c, Representative flow plot of NK cells isolated from lungs of Ctrl and anti-ASGM1 antibody treated mice (n = 3 mice). d, Total lung metastasis lesion numbers of Ctrl, anti-CSF-1R or anti-asialo GM1-treated mice (n = 15 mice per group). e, Treatment schematic and primary tumor weights of Ctrl and anti-NK1.1 antibody treated mice, or CD1d-KO mice at removal. f, Representative flow plot of depletion of blood NK cells by anti-NK1.1 antibody (n = 3 mice). g, Total lung metastasis lesion numbers of wildtype ctrl antibody-treated, wildtype anti-NK1.1-treated, or CD1d-KO mice. (For e, g, Ctrl, n = 13 mice, anti-NK1.1, n = 14 mice, CD1d KO, n = 12 mice). Mean ± s.d. (a, d, e, g) shown. P values were calculated by two-tailed unpaired Student’s t-test (a, e), two-tailed unpaired Mann–Whitney U-test (d, g).
Extended Data Fig. 4 Depletion of macrophages and T cells does not selectively increase monoclonal metastasis.
a, Treatment schematic and representative flow plots of lung-resident macrophages from bronchoalveolar lavage fluid of Ctrl liposome or clodrosome-treated mice (n = 3 mice). b, Lung metastasis growth curve and representative whole body mouse bioluminescence images of mice injected with single cells or clusters of AT3 cells following intranasal treatment with ctrl liposome or clodrosome (n = 5 mice). c, Treatment schematic and representative flow plot of blood T cells from Ctrl, anti-CD4 and anti-CD8 antibody treated mice. (n = 3 mice). d, e, Quantification of total lesions number and single/ mixed-colored lung metastases from wildtype mice treated with control, anti-CD4, or anti-CD8 antibody as absolute lesions counts (left and middle) or percentages of total (right) (Ctrl, n = 9; anti-CD4, n = 10; anti-CD8, n = 10 mice). Mean ± s.d. d, e, or s.e.m. (b) shown. P values were calculated by two-tailed unpaired Mann–Whitney U-test (d, e) or two-way repeated measures ANOVA (b).
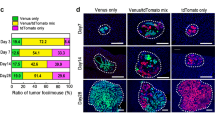
Extended Data Fig. 5 NK cell depletion attenuates the metastatic advantage of disseminated tumor cell clusters.
a, Treatment schematic and representative flow plots of blood NK cells in Rag2yc-dKO mice following NK cell adoptive transfer (Representative of n = 3 mice). b, Primary tumor volume of Rag2yc-dKO mice with or without NK cell adoptive transfer. c, Quantification and representative images of lung bioluminescence (as estimate of tumor burden) in Rag2yc-dKO mice following NK cell adoptive transfer (For b, c, Rag2yc-dKO, n = 11 mice; Rag2yc-dKO + NK, n = 10 mice). d, e, Lung metastasis growth curve and representative whole body mouse bioluminescence images of mice injected with single cells or clusters of e) AT3 cells and f) 4T1 cells following treatment with ctrl antibody or anti-asialo GM1 (n = 5 per group). (Mean ± s.d.(b, c) or s.e.m. d, e, shown. P values were calculated by two-tailed unpaired Student’s t-test (b), two-tailed unpaired Mann–Whitney U-test (c), or two-way repeated measures ANOVA.
Extended Data Fig. 6 Characterization of cluster-forming and non cluster-forming mouse breast cancer cell lines.
a, Representative microscopy images of cluster-forming and non cluster-forming cell lines in suspension following EDTA digestion. (n = 5 images/ cell line) Scale bar, 200 µm. b, Representative microscopy images of cluster-forming and non cluster-forming cells (GFP-labelled) cultured with or without NK cells for 4 hours. Scale bar, 200 µm (n = 3 independent experiments). c, Gene ontology pathways enriched in cluster-forming cell lines relative to non cluster-forming cell lines. P-values were calculated by hypergeometric tests and adjusted for multiple testing using FDR. (c).
Extended Data Fig. 7 Epithelial-to-mesenchymal transition alters cluster formation and expression of NK cell ligands.
a,b, Zeb1 expression levels (measured by qRT-PCR) and representative microscopy images of AT3 ctrl and miR-200c cells, T11 ctrl and miR-200c cells and PyMT-M and -E cells. (AT3, PyMT, n = 3; T11, n = 5 independent experiments) Scale bar, 100 µm. c, In vitro cell proliferation assay showing relative areas of T11 Ctrl and T11 miR-200c, and AT3 Ctrl and AT3 miR-200c. (n = 3 independent experiments). d, Representative microscopy images showing the invasive edge of AT3 tumor organoids from Ctrl and miR-200c cells cultured on Matrigel. Arrowheads show invasive single cells and arrows show collectively migrating clusters. (n = 5 organoids) Scale bar, 50 µm. e, Representative microscopy images of T11 ctrl and T11 miR-200c cells in suspension. Scale bar, 100 µm (n = 5 images/cell line). f, (Left) Gating strategy and histogram of flow cytometry analysis of Rae1 in T11 miR-200c, AT3 miR-200c, PyMT-E and PyMT-M cell lines. (Right) The fold change in Rae1 median fluorescent intensity (MFI) of PyMT-E cells over PyMT-M is shown. (n = 3 independent experiments). Mean ± s.e.m. shown. P values were calculated by two-tailed paired Student’s t-test (a, paired by batch), two-way repeated measures ANOVA (c), or one sample t test (f). N/A, not applicable.
Extended Data Fig. 8 Correlations between EMT characteristics, expression of NK cell ligands and cancer cell clustering in pre-clinical and clinical data.
a, EMT and NK cell activating gene signature scores calculated based on RNA-seq data of non cluster-forming and cluster-forming breast cancer cell lines (from GSE104765, n = 3 technical replicates/ cell line). b, (left) The correlation between NK cell activating score, and observed NK cell-mediated killing of cancer cells in vitro (right) Correlation between NK cell activating score and EMT score of cluster-forming and non cluster-forming cell lines (GSE104765, n = 3 technical replicates/ cell line). c, EMT and NK cell activating score of MCF10A cells with or without TGF-β treatment (from GSE81955, n = 3 independent experiments). d, EMT score of single and cluster CTCs isolated from breast cancer patients (from GSE51827, n = 15 pools of single CTCs and 14 CTC-clusters isolated from 10 breast cancer patients). (Mean ± s.e.m. shown. Pearson’s correlation coefficient was shown (b). P values were calculated by two-tailed unpaired Student’s t-test (a,c,d). For boxplots (a, d), the center shows the median; Upper and lower hinges show the first and third quartiles; and the upper and lower whiskers show highest and lowest values within 1.5 × IQR (interquartile range) of the hinge.
Extended Data Fig. 9 Altering epithelial phenotypes of tumor cells affects susceptibility to NK cell killing in vitro and in vivo.
a, Representative microscopy images from in vitro NK cell cytotoxicity assay of T11 Ctrl and T11 miR-200c, and AT3 Ctrl and AT3 miR-200c (n = 3 independent experiments/ cell line). b, Flow cytometric quantification of CD107a and IFN-y expression in NK cells co-cultured with PyMT-M and PyMT-E cells (n = 3 independent experiments). c, Flow cytometric gating strategy and representative flow plots showing quantification for CD107a and IFNγ expression in NK cells co-cultured with AT3 control and AT3 mir200c. d, Treatment schematic and lung metastasis (bioluminescence) growth curves of T11 ctrl and T11 miR-200c cells injected into the tail vein of control or anti-NKG2D antibody treated mice. (n = 5 mice per group). e, Metastatic burden from d) for Day 7. The log2 fold change in bioluminescence between miR200c-induced and uninduced is shown. Representative images are also shown (n = 5 mice per group). Mean ± s.e.m. shown. P values were calculated by two-tailed unpaired Student’s t-test (b, e) or repeated measures ANOVA (d).
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Lo, H.C., Xu, Z., Kim, I.S. et al. Resistance to natural killer cell immunosurveillance confers a selective advantage to polyclonal metastasis. Nat Cancer 1, 709–722 (2020). https://doi.org/10.1038/s43018-020-0068-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0068-9
This article is cited by
-
Splenectomy has opposite effects on the growth of primary compared with metastatic tumors in a murine colon cancer model
Scientific Reports (2024)
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
UFObow: A single-wavelength excitable Brainbow for simultaneous multicolor ex-vivo and in-vivo imaging of mammalian cells
Communications Biology (2024)
-
Research progress on the multi-omics and survival status of circulating tumor cells
Clinical and Experimental Medicine (2024)
-
Survival mechanisms of circulating tumor cells and their implications for cancer treatment
Cancer and Metastasis Reviews (2024)