Abstract
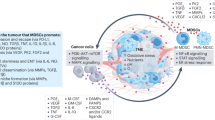
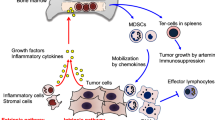
Immunotherapy that targets lymphoid cell checkpoints holds great promise for curing cancer. However, many cancer patients do not respond to this form of therapy. In addition to lymphoid cells, myeloid cells play essential roles in controlling immunity to cancer. Whether myeloid checkpoints exist that can be targeted to treat cancer is not well established. Here we show that c-Rel, a member of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) family, specified the generation of myeloid-derived suppressor cells by selectively turning on protumoral genes while switching off antitumoral genes through a c-Rel enhanceosome. c-Rel deficiency in myeloid cells markedly inhibited cancer growth in mice and pharmaceutical inhibition of c-Rel had the same effect. Combination therapy that blocked both c-Rel and the lymphoid checkpoint protein programmed cell death protein 1 was more effective in treating cancer than blocking either alone. Thus, c-Rel is a myeloid checkpoint that can be targeted for treating cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Microarray and RNA-seq data that support the findings of this study have been deposited in the ArrayExpress under accession nos. E-MTAB-8674 and E-MTAB-8714. Source data for Fig. 1 and Figs. 4–7 are provided with this paper and are available online. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 19, 133–150 (2019).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Kumar, V., Patel, S., Tcyganov, E. & Gabrilovich, D. I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 37, 208–220 (2016).
Manjili, M. H. Phenotypic plasticity of MDSC in cancers. Immunol. Invest. 41, 711–721 (2012).
Solito, S., Pinton, L., Damuzzo, V. & Mandruzzato, S. Highlights on molecular mechanisms of MDSC-mediated immune suppression: paving the way for new working hypotheses. Immunol. Invest. 41, 722–737 (2012).
Trikha, P. & Carson, W. E. 3rd Signaling pathways involved in MDSC regulation. Biochim. Biophys. Acta 1846, 55–65 (2014).
Bronte, V. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016).
Srivastava, M. K. et al. Myeloid suppressor cell depletion augments antitumor activity in lung cancer. PLoS ONE 7, e40677 (2012).
Stromnes, I. M. et al. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut 63, 1769–1781 (2014).
Steinberg, S. M. et al. Myeloid cells that impair immunotherapy are restored in melanomas with acquired resistance to BRAF inhibitors. Cancer Res. 77, 1599–1610 (2017).
Enciso-Mora, V. et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat. Genet. 42, 1126–1130 (2010).
Trynka, G. et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-κB signalling. Gut 58, 1078–1083 (2009).
Gregersen, P. K. et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 41, 820–823 (2009).
Hussman, J. P. et al. GWAS analysis implicates NF-κB-mediated induction of inflammatory T cells in multiple sclerosis. Genes Immun. 17, 305–312 (2016).
Himmelstein, D. S. & Baranzini, S. E. Heterogeneous network edge prediction: a data integration approach to prioritize disease-associated genes. PLoS Comput. Biol. 11, e1004259 (2015).
Beecham, A. H. et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013).
Baranzini, S, E. et al. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am. J. Hum. Genet. 92, 854–865 (2013).
Simek, S. & Rice, N. R. Detection and characterization of the protein encoded by the chicken c-rel protooncogene. Oncogene Res. 2, 103–119 (1988).
Grumont, R. J. & Gerondakis, S. The murine c-rel proto-oncogene encodes two mRNAs the expression of which is modulated by lymphoid stimuli. Oncogene Res. 5, 245–254 (1990).
Artis, D. et al. Differential requirement for NF-κB family members in control of helminth infection and intestinal inflammation. J. Immunol. 169, 4481–4487 (2002).
Carrasco, D. et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J. Exp. Med. 187, 973–984 (1998).
Reinhard, K. et al. c-Rel promotes type 1 and type 17 immune responses during Leishmania major infection. Eur. J. Immunol. 41, 1388–1398 (2011).
Harling-McNabb, L. et al. Mice lacking the transcription factor subunit Rel can clear an influenza infection and have functional anti-viral cytotoxic T cells but do not develop an optimal antibody response. Int. Immunol. 11, 1431–1439 (1999).
Liou, H. C. et al. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int. Immunol. 11, 361–371 (1999).
Köntgen, F. et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9, 1965–1977 (1995).
Campbell, I. K., Gerondakis, S., O’Donnell, K. & Wicks, I. P. Distinct roles for the NF-κB1 (p50) and c-Rel transcription factors in inflammatory arthritis. J. Clin. Invest. 105, 1799–1806 (2000).
Hilliard, B. A. et al. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J. Clin. Invest. 110, 843–850 (2002).
Lamhamedi-Cherradi, S.-E. et al. Transcriptional regulation of type I diabetes by NF-κB. J. Immunol. 171, 4886–4892 (2003).
Wang, Y. et al. c-Rel is essential for the development of innate and T cell-induced colitis. J. Immunol. 180, 8118–8125 (2008).
Jordan, K. A., Dupont, C. D., Tait, E. D., Liou, H.-C. & Hunter, C. A. Role of the NF-κB transcription factor c-Rel in the generation of CD8+ T-cell responses to Toxoplasma gondii. Int. Immunol. 22, 851–861 (2010).
Ruan, Q. et al. Development of Foxp3+ regulatory T cells is driven by the c-Rel enhanceosome. Immunity 31, 932–940 (2009).
Oh, H. An NF-κB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity 47, 450–465.e5 (2017).
Grinberg-Bleyer, Y. et al. NF-κB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell 170, 1096–1108.e13 (2017).
Ruan, Q. et al. The Th17 immune response is controlled by the Rel–RORγ–RORγT transcriptional axis. J. Exp. Med. 208, 2321–2333 (2011).
Youn, J. I., Collazo, M., Shalova, I. N., Biswas, S. K. & Gabrilovich, D. I. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 91, 167–181 (2012).
Gato-Cañas, M. et al. A core of kinase-regulated interactomes defines the neoplastic MDSC lineage. Oncotarget 6, 27160–27175 (2015).
Chen Y. H., Murali R. & Sun J. Rel inhibitors and methods of use thereof. US patent 8609730 (PCT/US2009/030325) (2009).
Marvel, D. & Gabrilovich, D. I. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest. 125, 3356–3364 (2015).
Marigo, I. et al. Tumor-induced tolerance and immune suppression depend on the C/EBPβ transcription factor. Immunity 32, 790–802 (2010).
Kaneda, M. M. et al. PI3Kγ is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016).
Ostrand-Rosenberg, S., Sinha, P., Beury, D. W. & Clements, V. K. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin. Cancer Biol. 22, 275–281 (2012).
Srivastava, M. K. et al. Targeting myeloid-derived suppressor cells augments antitumor activity against lung cancer. Immunotargets Ther. 2012, 7–12 (2012).
Zilio, S. & Serafini, P. Neutrophils and granulocytic MDSC: the Janus god of cancer immunotherapy. Vaccines (Basel) 4, 31 (2016).
Zhao, Y., Wu, T., Shao, S., Shi, B. & Zhao, Y. Phenotype, development, and biological function of myeloid-derived suppressor cells. Oncoimmunology 5, e1004983 (2016).
Knopp, M. M., Löbmann, K., Elder, D. P., Rades, T. & Holm, R. Recent advances and potential applications of modulated differential scanning calorimetry (mDSC) in drug development. Eur. J. Pharm. Sci. 87, 164–173 (2016).
Merika, M. & Thanos, D. Enhanceosomes. Curr. Opin. Genet. Dev. 11, 205–208 (2001).
Panne, D. The enhanceosome. Curr. Opin. Struct. Biol. 18, 236–242 (2008).
Arnosti, D. N. & Kulkarni, M. M. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890–898 (2005).
Tumang, J. R. et al. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur. J. Immunol. 28, 4299–4312 (1998).
Acknowledgements
We thank H.-C. Liou (Cornell University), W. Weinberg (US FDA) and J. Zakrzewski (Cornell University) for providing the breeding pair of the Rel−/− mice. We thank D. Gabrilovich, J. Goldsmith, P. Fang, L. Wan, M. Lin, D. Zhang, L. Guan, J. Devergiilis and J. Sun for valuable discussions, technical support and reagents. We thank the University of Pennsylvania Pancreatic Islet Cell Biology Core and D.P. Beiting from the University of Pennsylvania School of Veterinary Medicine for technical support. This work was supported in part by grants from the National Institutes of Health (nos. R01-AI152195, R01-AI099216, R01-AI121166, R01-AI143676 and R01-AI136945 to Y.H.C.); X.L. was partially supported by grant no. NIH-T32-DK007780.
Author information
Authors and Affiliations
Contributions
T.L. and X.L. designed and executed the experiments and wrote the manuscript. A.Z. designed and performed some of the MDSC suppression experiments. H.S., M.L., W.W., C.-N.L., G.L., E.E., Q.R. and W.S. helped complete certain molecular, cellular or animal experiments. S.G. provided the Rel gene floxed mice. J.J. and R.M. designed and optimized the c-Rel inhibitor. Y.H.C. conceived and supervised the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.H.C. and R.M. are inventors of the following patent that describes the c-Rel inhibitor used in this study: Chen Y.H., Murali R. & Sun J. Rel inhibitors and methods of use thereof (USA patent no. US8609730B2). Y.H.C. is a member of the advisory board of Amshenn Pharmaceutical Company and Binde Company.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Global and myeloid Rel gene deletion blocks tumor growth and reduces MDSCs in mice.
a, Tumor growth in WT and Rel−/− mice (n = 5 mice/group) injected s.c. with B16F0 tumor cells (***, P < 0.0001). b, Tumor weight of WT and Rel−/− mice (n = 10 mice/group) treated in Fig. 1a. (*, P = 0.0188). c, Percentages of CD11b+Gr-1+ cells in the blood of WT and Rel−/− mice treated with anti-Gr-1 or IgG isotype control (n = 3 mice/group), 7 days post B16F10 tumor cell inoculation (**, P = 0.0037; ***, P < 0.0001). d, Percentages of CD11b+Gr-1+ cells in the tumor of WT mice treated with anti-Gr-1 or IgG isotype control (n = 3 mice/group), 7 days post B16F10 tumor cell inoculation (***, P = 0.0008). e, Percentages of CD4+CD25+ cells in the spleen of WT and Rel−/− mice treated with anti-CD25 or IgG isotype control (n = 3 mice/group), 14 days post B16F10 tumor cell inoculation (***, P < 0.0001). f, Tumor size on Day 14 of WT and Rel−/− mice treated as in Fig. 1c–e. n = 16 for the WT + IgG group, n = 15 for the Rel−/−+IgG group, n = 11 for the WT + anti-Gr1 group, n = 10 for the Rel−/−+anti-Gr1 group, n = 9 for the WT + anti-CD25 group, and n = 9 for the Rel−/−+anti-CD25 group (*, P = 0.0303; **, P < 0.01; ***, P = 0.0001). g, c-Rel expression in Gr-1+ and Gr-1- splenocytes of LyzM-Cre (Cre) and LyzM-Cre RelF/F (RelF/F) tumor-bearing mice as determined by Western blot. Representative blots from biologically independent experiments were shown and the bar graph shows the relative quantities of the c-Rel protein in the corresponding group shown below (n = 3 mice for each group; *, P = 0.0101). h,i, Percentages of CD11b+Ly6G+ (h) and CD11b+Ly6C+ (i) leukocytes in the tumor of WT and Rel−/− mice treated in Fig. 1a (n = 5 mice/group). **, P = 0.0011 for panel h; **, P = 0.0087 for panel i. j, Percentages of CD8+CD25+ leukocytes in the tumor of LyzM-Cre and LyzM-Cre RelF/F mice treated in Fig. 1f (n = 8 mice/group; ***, P = 0.0005). k-n, Percentages of the indicated leukocyte subsets in the spleen of WT and Rel−/− mice treated in Fig. 1a. (n = 4 mice/group in the panels k and l; n = 5 mice/group in the panels m and n; *, P = 0.037). Statistical significance was determined by two-tailed Mann-Whitney U-test (a), two-tailed unpaired t-test (b-f, h-j, m), or one-way ANOVA with Tukey post-hoc test (g). For all panels, data are presented as means ± s.e.m.
Extended Data Fig. 2 Percentages of immune cell subsets in tumor-bearing and naïve LyzM-Cre and LyzM-Cre RelF/F mice.
a, Percentages of CD11b+Gr-1+ cells in the spleen of mice treated in Fig. 1f. n = 6 mice/group. b-d, Percentages of CD4+ cells in the spleen (b, n = 6/group), blood (c, n = 6/group), and tumor (d, n = 8/group) of mice treated in Fig. 1f. e-g, Percentages of CD4+CD25+ cells in the spleen (e, n = 6/group), blood (f, n = 6 for the LyzM-Cre group and n = 5 for the LyzM-Cre RelF/F group), and tumor (g, n = 3 for the LyzM-Cre group and n = 6 for the LyzM-Cre RelF/F group) of mice treated in Fig. 1f. h-j, Percentages of CD8+ cells in the spleen (h, n = 6/group), blood (i, n = 6/group), and tumor (j, n = 8/group) of mice treated in Fig. 1f. k-n, Percentages of the indicated leukocyte subsets in the tumor (n = 3 for the LyzM-Cre group and n = 6 for the LyzM-Cre RelF/F group) of mice treated in Fig. 1f (k, l), and the spleen (n = 3 for the LyzM-Cre group and n = 5 for the LyzM-Cre RelF/F group) of naïve mice (m, n). (***, P = 0.0002) Statistical significance was determined by two-tailed unpaired t-test (k). For all panels, data are presented as means ± s.e.m.
Extended Data Fig. 3 Reduced suppressive function, reactive oxygen species (ROS) production, and cell growth in Rel−/− MDSCs.
a,b, Representative flow cytometry plots from the MDSC-T cell suppression assay for Fig. 2a (a) and Fig. 2b (b). c, Growth of bone marrow-derived MDSCs from WT (n = 4 mice/group) and Rel−/− (n = 9 mice/group) mice. (*, P = 0.03299). d, ROS production by bone marrow-derived MDSCs from WT and Rel−/− mice (n = 6 mice/group; ***, P < 0.0001). e, Percentages of CD11b+Ly6G+ and CD11b+Ly6C+ cells in bone marrow-derived MDSCs from WT and Rel−/− mice. Data representative of three independent experiments. f, Tumor growth in WT mice injected s.c. with B16F10 tumor cells plus Rel−/− MDSCs infected with control (n = 5 mice) or Rel-expressing retroviruses (n = 6 mice) (**, P = 0.0041). g, Percentages of CD44high cells in intratumor CD8+ cells of WT mice treated as in panel f. (n = 4 mice/group; *, P = 0.0187). h, The degree of suppression, at the indicated Effector:T cell ratio, of CD8+ T cell proliferation by bone marrow-derived MDSCs and BMDMs from naïve mice (n = 3 mice/group). The concentrations of anti-CD3 and anti-CD28 used (125 ng/mL each) were half of those in Fig. 2b (***, P < 0.0001). i, Representative flow cytometry plots for the MDSC-T cell suppression assay for Panel h. Statistical significance was determined by two-tailed unpaired t-test (c-e, g), two-tailed Mann-Whitney U-test (f), or two-way ANOVA with Tukey post-hoc test (h). Data are presented as means ± s.e.m. (c,d,f,h).
Extended Data Fig. 4 Rel gene deletion in MDSCs leads to upregulated expression of inflammatory genes and decreased Cebpb downstream genes.
a,b, Results from Ingenuity Pathway analysis of the RNA-seq data of bone marrow-derived WT and Rel−/− MDSCs, showing upregulated ‘inflammatory response’ genes (a, red) and downregulated Cebpb downstream genes (b, green) in Rel−/− MDSCs. Statistical significance was determined by calculated a right-tailed Fisher’s Exact Test. All data are pooled from 3 independent experiments.
Extended Data Fig. 5 Rel gene deletion in macrophages results in decreased expression of inflammatory genes.
BMDMs from WT and Rel−/− mice were treated with vehicle or LPS (100 ng/mL) for 1 hour and gene microarray analysis was performed for ~30,000 murine genes. Ingenuity Pathway Analysis was performed to identify c-Rel-regulated genes that were downstream of LPS response. Blue genes were decreased, and red genes were increased in Rel−/− cells as compared to WT cells, after LPS treatment.
Extended Data Fig. 6 The inhibitor is c-Rel-specific and blocks MDSC suppressive functions.
a, Relative numbers of WT and Rel−/− human Jurkat T cells treated with the c-Rel inhibitor (2.5 μM) or vehicle for the indicated times (n = 3 biologically independent cultures/group). b,c, Relative numbers of EL4 (b, n = 3 biologically independent cultures/group) and B16F10 (c, n = 3 biologically independent cultures/group) cells treated with or without the c-Rel inhibitor (5 μM) (**, P = 0.00767). d,e, Relative mRNA levels of IL-2 in WT, Rel−/− human Jurkat T cells (d, n = 3 biologically independent cultures/group; **, P = 0.0047), and normal human PBMCs (e, n = 3 biologically independent cultures/group; *, P = 0.0241) that were treated with or without c-Rel inhibitor (1 μM). For stimulation, plate-coated anti-mouse CD3 (250 ng/ml) plus soluble anti-mouse CD28 (250 ng/ml), or PMA (10 ng/ml) plus ionomycin (1 µM) were added to the culture for 4 hours, as indicated. f, Preferential inhibition of c-Rel binding to DNA by the c-Rel inhibitor. Western blot for the indicated NF-κB proteins after the NF-κB oligonucleotide pull-down of the nuclear extracts of WT and Rel−/− splenocytes treated with the c-Rel inhibitor (5 µM) or vehicle control (Ctr). g, Body weight change of mice that were injected i.p. with vehicle control only (n = 7 mice), or injected s.c. with B16F10 cells and i.p. with the c-Rel inhibitor (n = 3 mice) or vehicle control (n = 4 mice) as indicated. h,i, Representative flow cytometry plots of the MDSC-T cell suppression assay for Fig. 6e (h) and Fig. 6f (i). Data are presented as means ± s.e.m (a-e, g, and i). n = 3 mice/group (d,e,j). Statistical significance was determined by two-tailed unpaired t-test (b-e).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and gels.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Li, T., Li, X., Zamani, A. et al. c-Rel is a myeloid checkpoint for cancer immunotherapy. Nat Cancer 1, 507–517 (2020). https://doi.org/10.1038/s43018-020-0061-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0061-3
This article is cited by
-
FFAR2 expressing myeloid-derived suppressor cells drive cancer immunoevasion
Journal of Hematology & Oncology (2024)
-
Myeloid-derived suppressor cells in cancer and cancer therapy
Nature Reviews Clinical Oncology (2024)
-
NF-κB in biology and targeted therapy: new insights and translational implications
Signal Transduction and Targeted Therapy (2024)
-
Aryl hydrocarbon receptor nuclear translocator limits the recruitment and function of regulatory neutrophils against colorectal cancer by regulating the gut microbiota
Journal of Experimental & Clinical Cancer Research (2023)
-
Regulatory cells and the effect of cancer immunotherapy
Molecular Cancer (2023)