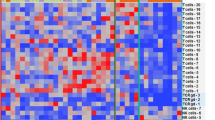
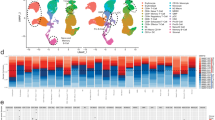
Abstract
Precursor states of multiple myeloma (MM) and its native tumor microenvironment need in-depth molecular characterization to better stratify and treat patients at risk. Using single-cell RNA sequencing of bone marrow cells from precursor stages, monoclonal gammopathy of unknown significance and smoldering MM, to full-blown MM alongside healthy donors, we demonstrate early immune changes during patient progression. We find that natural killer cell abundance is frequently increased in the early stages and associated with altered chemokine receptor expression. As early as smoldering MM, we show loss of granzyme K+ memory cytotoxic T cells and show their critical role in MM immunosurveillance in mouse models. Finally, we report major histocompatibility complex class II dysregulation in CD14+ monocytes, which results in T-cell suppression in vitro. These results provide a comprehensive map of immune changes at play over the evolution of premalignant MM, which will help develop strategies for immune-based patient stratification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The scRNA-seq data that support the findings of this study have been deposited with the Gene Expression Omnibus under accession code GSE124310. Raw sequencing data have been deposited with the database of Genotypes and Phenotypes (phs001323.v1.p1). Previously published microarray data that were reanalyzed in this study are available under accession code GSE6477. Source data for Figs. 1–6 and Extended Data Figs. 2–4 and 6–9 have been provided in corresponding Source Data tables. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The single-cell RNA data was processed using cellranger v.2.0.1 (https://www.10xgenomics.com/) and analyzed with the R package Seurat v.2.3.1 (https://satijalab.org/seurat/). Gene expression signatures were extracted using our SignatureAnalyzer algorithm available on github (https://github.com/broadinstitute/getzlab-SignatureAnalyzer).
References
Kyle, R. A. & Rajkumar, S. V. Multiple myeloma. Blood 111, 2962–2972 (2008).
Landgren, O. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: biological insights and early treatment strategies. Hematology Am. Soc. Hematol. Educ. Program 2013, 478–487 (2013).
Rajkumar, S. V. Multiple myeloma: 2011 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 86, 57–65 (2011).
Kyle, R. A. et al. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia 24, 1121–1127 (2010).
Rajkumar, S. V. et al. Serum free light chain ratio is an independent risk factor for progression in monoclonal gammopathy of undetermined significance. Blood 106, 812–817 (2005).
Rajkumar, S. V. et al. Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia 27, 1738–1744 (2013).
Hanamura, I. et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 108, 1724–1732 (2006).
Manier, S. et al. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 14, 100–113 (2017).
Bladé, J., Dimopoulos, M., Rosiñol, L., Rajkumar, S. V. & Kyle, R. A. Smoldering (asymptomatic) multiple myeloma: current diagnostic criteria, new predictors of outcome, and follow-up recommendations. J. Clin. Oncol. 28, 690–697 (2010).
Kyle, R. A. et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N. Engl. J. Med. 356, 2582–2590 (2007).
Landgren, O. et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 113, 5412–5417 (2009).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Tyekucheva, S. et al. Stromal and epithelial transcriptional map of initiation progression and metastatic potential of human prostate cancer. Nat. Commun. 8, 420 (2017).
Glavey, S. V. et al. Proteomic characterization of human multiple myeloma bone marrow extracellular matrix. Leukemia 31, 2426–2434 (2017).
El-Haibi, C. P. et al. Critical role for lysyl oxidase in mesenchymal stem cell-driven breast cancer malignancy. Proc. Natl Acad. Sci. USA 109, 17460–17465 (2012).
Pearce, O. M. T. et al. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. 8, 304–319 (2018).
Finak, G. et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 14, 518–527 (2008).
Calon, A. et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47, 320–329 (2015).
Petitprez, F. et al. PD-L1 expression and CD8+ T-cell infiltrate are associated with clinical progression in patients with node-positive prostate cancer. Eur. Urol. Focus 5, 192–196 (2019).
Zhang, L. et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 348, 203–213 (2003).
Podar, K., Hideshima, T., Chauhan, D. & Anderson, K. C. Targeting signalling pathways for the treatment of multiple myeloma. Expert Opin. Ther. Targets 9, 359–381 (2005).
Klein, B. Cytokine, cytokine receptors, transduction signals, and oncogenes in human multiple myeloma. Semin. Hematol. 32, 4–19 (1995).
Sawanobori, M. et al. Natural killer cell frequency and serum cytokine levels in monoclonal gammopathies: correlation of bone marrow granular lymphocytes to prognosis. Acta Haematol. 98, 150–154 (1997).
Pessoa de Magalhães, R. J. et al. Analysis of the immune system of multiple myeloma patients achieving long-term disease control by multidimensional flow cytometry. Haematologica 98, 79–86 (2013).
Bailur, J. K. et al. Early alterations in stem-like/resident T cells, innate and myeloid cells in the bone marrow in preneoplastic gammopathy. JCI Insight 4, e127807 (2019).
Leone, P. et al. Dendritic cells accumulate in the bone marrow of myeloma patients where they protect tumor plasma cells from CD8+ T-cell killing. Blood 126, 1443–1451 (2015).
Kawano, Y. et al. Blocking IFNAR1 inhibits multiple myeloma-driven Treg expansion and immunosuppression. J. Clin. Invest. 128, 2487–2499 (2018).
Feng, X. et al. Targeting CD38 suppresses induction and function of T regulatory cells to mitigate immunosuppression in multiple myeloma. Clin. Cancer Res. 23, 4290–4300 (2017).
Ramachandran, I. R. et al. Myeloid-derived suppressor cells regulate growth of multiple myeloma by inhibiting T cells in bone marrow. J. Immunol. 190, 3815–3823 (2013).
Nakamura, K. et al. Dysregulated IL-18 Is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell 33, 634–648.e5 (2018).
Zheng, Y. et al. Macrophages are an abundant component of myeloma microenvironment and protect myeloma cells from chemotherapy drug-induced apoptosis. Blood 114, 3625–3628 (2009).
De Beule, N. et al. Tumour-associated macrophage-mediated survival of myeloma cells through STAT3 activation. J. Pathol. 241, 534–546 (2017).
Osterborg, A., Nilsson, B., Björkholm, M., Holm, G. & Mellstedt, H. Natural killer cell activity in monoclonal gammopathies: relation to disease activity. Eur. J. Haematol. 45, 153–157 (1990).
Scavelli, C. et al. Vasculogenic mimicry by bone marrow macrophages in patients with multiple myeloma. Oncogene 27, 663–674 (2008).
Minnie, S. A. et al. Myeloma escape after stem cell transplantation is a consequence of T-cell exhaustion and is prevented by TIGIT blockade. Blood 132, 1675–1688 (2018).
Dispenzieri, A. et al. Immunoglobulin free light chain ratio is an independent risk factor for progression of smoldering (asymptomatic) multiple myeloma. Blood 111, 785–789 (2008).
Perussia, B. Fc receptors on natural killer cells. Curr. Top. Microbiol. Immunol. 230, 63–88 (1998).
Yang, C. et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 10, 3931 (2019).
Bernardini, G., Sciumè, G. & Santoni, A. Differential chemotactic receptor requirements for NK cell subset trafficking into bone marrow. Front. Immunol. 4, 12 (2013).
Baecher-Allan, C., Wolf, E. & Hafler, D. A. MHC class II expression identifies functionally distinct human regulatory T cells. J. Immunol. 176, 4622–4631 (2006).
Karalis, K. et al. Autocrine or paracrine inflammatory actions of corticotropin-releasing hormone in vivo. Science 254, 421–423 (1991).
Bratke, K., Kuepper, M., Bade, B., Virchow, J. C. Jr. & Luttmann, W. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol. 35, 2608–2616 (2005).
Ahn, E. et al. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl Acad. Sci. USA 115, 4749–4754 (2018).
Odorizzi, P. M., Pauken, K. E., Paley, M. A., Sharpe, A. & Wherry, E. J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137 (2015).
Roccaro, A. M. et al. CXCR4 regulates extra-medullary myeloma through epithelial-mesenchymal-transition-like transcriptional activation. Cell Rep. 12, 622–635 (2015).
Brown, R. D. et al. Prognostically significant cytotoxic T cell clones are stimulated after thalidomide therapy in patients with multiple myeloma. Leuk. Lymphoma 50, 1860–1864 (2009).
Raitakari, M. et al. T-cell expansions in patients with multiple myeloma have a phenotype of cytotoxic T cells. Br. J. Haematol. 110, 203–209 (2000).
Tienhaara, A. & Pelliniemi, T. T. Peripheral blood lymphocyte subsets in multiple myeloma and monoclonal gammopathy of undetermined significance. Clin. Lab. Haematol. 16, 213–223 (1994).
Wu, L. et al. Lenalidomide enhances antibody-dependent cellular cytotoxicity of solid tumor cells in vitro: influence of host immune and tumor markers. Cancer Immunol. Immunother. 60, 61–73 (2011).
Jurisic, V., Srdic, T., Konjevic, G., Markovic, O. & Colovic, M. Clinical stage-depending decrease of NK cell activity in multiple myeloma patients. Med. Oncol. 24, 312–317 (2007).
Ponzetta, A. et al. Multiple myeloma impairs bone marrow localization of effector natural killer cells by altering the chemokine microenvironment. Cancer Res. 75, 4766–4777 (2015).
Zelle-Rieser, C. et al. T cells in multiple myeloma display features of exhaustion and senescence at the tumor site. J. Hematol. Oncol. 9, 116 (2016).
Castella, B., Foglietta, M., Riganti, C. & Massaia, M. Vγ9Vδ2 T Cells in the bone marrow of myeloma patients: a paradigm of microenvironment-induced immune suppression. Front. Immunol. 9, 1492 (2018).
Chan, A. C. et al. Natural killer T cell defects in multiple myeloma and the impact of lenalidomide therapy. Clin. Exp. Immunol. 175, 49–58 (2014).
Dhodapkar, M. V. & Richter, J. Harnessing natural killer T (NKT) cells in human myeloma: progress and challenges. Clin. Immunol. 140, 160–166 (2011).
Castella, B., Melaccio, A., Foglietta, M., Riganti, C. & Massaia, M. Vγ9Vδ2 T cells as strategic weapons to improve the potency of immune checkpoint blockade and immune interventions in human myeloma. Front. Oncol. 8, 508 (2018).
Ortiz, M. L., Lu, L., Ramachandran, I. & Gabrilovich, D. I. Myeloid-derived suppressor cells in the development of lung cancer. Cancer Immunol. Res. 2, 50–58 (2014).
Fang, Z. et al. Myeloid-derived suppressor cell and macrophage exert distinct angiogenic and immunosuppressive effects in breast cancer. Oncotarget 8, 54173–54186 (2017).
Gabrilovich, D. I., Ostrand-Rosenberg, S. & Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 12, 253–268 (2012).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Taylor-Weiner, A. et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 20, 228 (2019).
Tan, V. Y. F. & Févotte, C. Automatic relevance determination in nonnegative matrix factorization with the β-divergence. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1592–1605 (2013).
Acknowledgements
We thank the Dana-Farber/Harvard Cancer Center in Boston for the use of the Specialized Histopathology Core for histology and immunohistochemistry service. The Dana-Farber/Harvard Cancer Center is supported in part by a National Cancer Institute Cancer Center Support Grant no. NIH 5 P30 CA06516. G.G. was partially funded by the Paul C. Zamecnik Chair in Oncology, Massachusetts General Hospital Cancer Center. N.J.H. was partially funded by G.G. startup funds at Massachusetts General Hospital, R01 (principal investigator I.M.G.) and the Harvard Graduate Program in Biophysics, Harvard University. Molecular Biophysics Training Grant NIH/NIGMS T32 GM008313 (N.J.H.; principal investigtor J. M. Hogle). This study was partially funded by two National Institutes of Health grants (no. NIH R01 CA 205954 and R01CA181683), the Leukemia and Lymphoma Society, the Multiple Myeloma Research Foundation and Stand Up to Cancer.
Author information
Authors and Affiliations
Contributions
O.Z., T.H.M., I.M.G., N.J.H. and G.G. conceived and designed the study. T.H.M. collected patient data and performed cytogenetic analyses. O.Z. generated the single-cell sequencing and CyTOF data, and performed in vivo experiments. O.Z. and T.H.M. performed in vitro experiments. N.J.H. processed and analyzed the single-cell sequencing data. O.Z. and N.J.H. integrated and interpreted the data. M.G., M.C., J.L.G. and S.A.M. assisted in setup of the study. J.P., M.X.H., C.-J.L., Y.E.M. and E.M.V.A. performed preliminary analyses. N.K.S., D.H. and B.B. managed clinical samples. R.F., E.B., B.F., A.F., S.C., M.P.A., M. Reidy, M. Rahmat, S.M. and M.B. performed validation experiments. I.M.G., G.G. and J.A. supervised the study. O.Z., N.J.H., T.H.M., R.S.P., G.G. and I.M.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect and POLYSOLVER. I.M.G. has a consulting/advisory role with GSK, AbbVie and Bristol Myers Squibb. I.M.G has a consulting role with Sanofi, Janssen Pharmaceuticals, Takeda, Celgene, Karyopharm, GNS, Cellectar Biosciences, Medscape, Genetech, Adaptive Biotechnologies, Aptitude, Curio Science, Magenta Therapeutics and Oncopeptides. I.M.G. received research funding/ honoraria from Celgene, Takeda, Bristol Myers Squibb, Janssen Pharmaceuticals and Amgen.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Marker genes demonstrating cell type identity of immune cell clusters.
tSNE representation of CD45 cell populations. In each subplot, cells are colored by log-normalized expression values for a given cell type specific marker gene.
Extended Data Fig. 2 Single-cell RNA sequencing results indicate changes in NK cell compartment as compared to healthy donors.
a, A significant increase in the fraction of NK cells in patients with malignant cells expressing IgG heavy chain. Fractions of NK cells are plotted for patients grouped by immunoglobulin heavy chain. Violin plots show minimum, median, and maximum values. A two-tailed t-test was performed on n=19 patient samples with df=12. b, Spearman correlation between NK cell fraction and CXCR4+ subset frequency calculated on 10,0000 samples with replacement of data points. 95% confidence interval is shown. c, Grouping of samples above and below median values for NK cell frequencies and CXCR4+ subset fractions. Points on the median were assigned in the conservative direction (that is to obtain a less significant p-value). A two-sided fisher-exact test was performed on n=23 patient samples.
Extended Data Fig. 3 Compositional alterations in immune populations of diseased patients as compared to healthy donors.
CyTOF profiling of 4 healthy donors and 13 MGUS-MM patients show a, significantly increased numbers of CD3+CD4+ T cells in BM aspirates of the patients as compared to healthy BM with mean values of 2.3 and 11.12 for CD3+CD4+, 1.6 and 3.7 for NK, and 1.5 and 0.5 for dendritic cells (DC) in healthy donors and MM patients respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent SD. b, Plasmacytoid DC (pDC) are significantly enriched in healthy BM as compared to MM patients with mean values of 1.2 and 0.26 respectively. Mean values for monocytic DC (mDC) are 3.7 and 3.2 for healthy donors and MM patients respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent average deviation. c, Classical monocytes (c Monoc) are enriched in BM aspirates of healthy donors compared to MM patients with respective mean values of 67.7 and 26.5. Mean values for non-classical monocytes (nc Monoc) were 13.1 and 44.4 for healthy donors and MM patients respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent average deviation. d, Proportion of CD66b+ cells in BM aspirates of SMM patients and healthy donors. Mean values are 23.8 and 20.6 for healthy donors and MM patients respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent SD.
Extended Data Fig. 4 Transcriptional alterations in cytotoxic T cell populations of diseased patients as compared to healthy donors.
a, Distribution of the Granzyme expression in T-cell cluster. GZMA+GZMB− corresponds to the Granzyme K expressing cells. b, Normalized expression values for four exhaustion-related genes across different T cell subsets, pooling cells across patients. c, Normalized expression values for four exhaustion-related genes across different donors and patients, pooling cells across T subsets. d, Heatmap of immune checkpoint molecules expression levels on different subsets of bone marrow T cells in SMM patients (n=8) as compared to healthy donors (n=4). Individual patients show increased levels of PD-1 and TIGIT in GrB expressing effector cells. Colored scale represents transformed ratio of protein expression. Barplots show variable expression of TIGIT (with mean values of 0.01 vs 0.12), PD-1 (0.52 vs. 0.55) and TIM-3 (0.39 vs. 0.51) in GrB-expressing T effectors from healthy BM and SMM patients respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent SD. e, Healthy memory cells show significantly higher expression of PD-1 compared to those of SMM patients with mean values of 19.5 and 4.7 respectively. Significant difference between groups was tested using a two-sided t-test. Bars represent SD.
Extended Data Fig. 5 IFN type-1 signaling increases in plasma cells during disease progression.
Expression of ISG-15 a, and MX1 b, increases during disease progression. Significant difference is observed between MGUS and MM stage, indicating an increase of IFN signals at later stages in the disease progression (GSE6477). Box plot displays the first quartile, median and third quartile for the gene expression levels, bars indicate the minimum and maximum value. Significant difference between groups was tested using one-way ANOVA and Tukey multiple comparison tests for healthy donors (ND, n=15), MGUS (n=21), SMM (n=23) and newly diagnosed MM (n=75).
Extended Data Fig. 6 Marker genes demonstrating cell type identity of monocytic clusters.
a, Mean expression of MHC II encoding genes. Violin plots show minimum, median, and maximum values. A BH-corrected two-tailed t-test was performed on n=32 patient samples. b, Heatmap of expression values for the top 10 genes with enriched expression in all monocytes discovered by k-nearest neighbors’ subclustering. Expression values are centered and normalized for each gene.
Extended Data Fig. 7 Dysregulation of HLA-DR surface representation in monocytes from diseased environment.
a, qPCR data demonstrate significant increase of HLA-DR expression in CD14+ monocytes after co-culture with MM cell lines. CD14+ cells alone and those co-cultured with B cells were used as a control. Median ratios were 1.54 for B cells, 2.07 for MM1.S cells, 3.28 for KMS-18, 8.34 for RPMI and 3.12 for OPM2 cells as compared to CD14+ control (1.0). Bars represent SD. Experiment was performed twice with independent donors in triplicates. Representative data from one experiment is shown. b, Immunofluorescence staining of tissue microarrays from MGUS, SMM and MM patients (n=45, TMA performed in triplicates, total of 135 BM sections analysed) demonstrates prevalent intracellular accumulation of HLA-DR (green) in CD14-expressing monocytes (red) in disease settings. Membrane-bound localization of HLA-DR was observed in healthy bone marrow monocytes. (yellow arrows point on cells with HLA-DR localized to the cell membrane, white arrows point on cells with HLA-DR accumulated in the cytoplasm.
Extended Data Fig. 8 MARCH-1 dependent internalization of HLA-DR in CD14+ monocytes in myeloma environment.
a, Gating strategy for HLA-DR on CD14+ cells. b, Knockdown of MARCH-1 by siRNA rescues presentation of HLA-DR molecules on the surface of CD14+ monocytes co-cultured with MM cells. Representative FACS profiles show higher numbers of HLA-DR+ cells after MARCH-1 knockdown with siRNA (experiment performed 3 times with 3 different donors using 2 different cell lines/2 different siRNA for MARCH-1, non-targeting si-RNA is used as a control). c, Mean fluorescence intensity demonstrates the two to 4.5-fold increase in the levels of HLA-DR protein expression on the cell surface of CD14+ cells. d, qPCR data for relative expression of MARCH-1 in CD14+ cells after siRNA knockdown as compared to the si-RNA control. The assay was performed twice with independent donors/2 cell lines/2 siRNAs, performed in triplicates. Representative data from one experiment is shown. In MM1.S cells, siRNA knockdown leads to reduction of MARCH-1 expression to 0.46 and 0.32 (median value, amplified with primer pair 2) and to 0.40 and 0.26 (median value, amplified with primer pair 3) as compared to control siRNA (median value, 1.0). In OPM2 cells, siRNA knockdown leads to reduction of MARCH-1 expression to 0.65 and 0.7 (median value, amplified with primer pair 2) and to 0.29 and 0.36 (median value, amplified with primer pair 3) as compared to control siRNA (median value, 1.0). Bars represent STDEVP. e, CD14+ cells with lower levels of MARCH-1 have increased HLA-DR protein on their cell surface. The experiment was performed twice with two independent donors/2 cell lines/2 siRNAs. f, Stronger correlation of DAPI and HLA-DR localization in MM1.S and OPM2 cells in control cells as compared to those after MARCH-1-siRNA transfection. The experiment was performed twice with two independent donors/2 cell lines/2 siRNAs.
Extended Data Fig. 9 CD14+ monocytes do not show the M-MDSC phenotype after co-culture with MM cells.
Expression of different markers for a, macrophages/monocytes and b, MDSCs on CD14+ cells in patients and healthy donors. c, CD14+ cells from healthy donors were co-cultured with MM cells. FACS analysis was performed on day 3 after co-culture. Representative results from one out of two independent experiments performed with two healthy donors/2 different cell lines. Due to restricted cell numbers, no replicates could be used. All donors have similar distribution of cells as compared to controls.
Extended Data Fig. 10 Alterations in tumor microenvironment start from the precursor stages of the MM and exhibit heterogeneous changes in the immune cell repertoire.
Illustration of immune alterations observed during progression. Bars begin at the stage in which they are first observed in our dataset.
Supplementary information
Supplementary Tables
Supplementary Table 1. Patient data and clinical information. List of clinical measurements for samples used for single-cell RNA sequencing. Supplementary Table 2. Sample information. Bead selection and quality metrics for single-cell RNA samples. Supplementary Table 3. Marker genes demonstrating cell type identity of immune cell clusters. List of top 10 differentially expressed genes with average log fold change (mean expression in within-cluster cells compared to mean expression of all out-of-cluster cells) and FDR-corrected rank-sum P values in immune cell clusters discovered by k-nearest neighbors clustering on cells from n = 32 patient samples.
Source data
Source Data Fig. 1
Data source. Primary data generated in experiments
Source Data Fig. 2
Data source. Primary data generated in experiments
Source Data Fig. 3
Data source. Primary data generated in experiments
Source Data Fig. 3
Unprocessed gel
Source Data Fig. 4
Data source. Primary data generated in experiments
Source Data Fig. 5
Data source. Primary data generated in experimens/repeats reported in the manuscript are provided
Source Data Fig. 6
Data source. Primary data generated in experimens/repeats reported in the manuscript are provided
Source Data Extended Data Fig. 2
Data source. Primary data generated in experiments
Source Data Extended Data Fig. 3
Data source. Primary data generated in experiments
Source Data Extended Data Fig. 4
Data source. Primary data generated in experiments
Source Data Extended Data Fig. 6
Data source. Primary data generated in experiments
Source Data Extended Data Fig. 7
Data source. Primary data generated in experiments
Source Data Extended Data Fig. 8
Data source. Primary data generated in experiments/repeats reported in the manuscript are provided
Source Data Extended Data Fig. 9
Data source. Primary data generated in experiments
Rights and permissions
About this article
Cite this article
Zavidij, O., Haradhvala, N.J., Mouhieddine, T.H. et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer 1, 493–506 (2020). https://doi.org/10.1038/s43018-020-0053-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0053-3
This article is cited by
-
The bone ecosystem facilitates multiple myeloma relapse and the evolution of heterogeneous drug resistant disease
Nature Communications (2024)
-
Bispecific BCMA/CD24 CAR-T cells control multiple myeloma growth
Nature Communications (2024)
-
Bone marrow inflammation in haematological malignancies
Nature Reviews Immunology (2024)
-
Single-cell technologies in multiple myeloma: new insights into disease pathogenesis and translational implications
Biomarker Research (2023)
-
Identification of potential resistance mechanisms and therapeutic targets for the relapse of BCMA CAR-T therapy in relapsed/refractory multiple myeloma through single-cell sequencing
Experimental Hematology & Oncology (2023)