Abstract
Alpelisib is a selective inhibitor of phosphoinositide 3-kinase (PI3K)α, shown to improve outcomes for PIK3CA-mutant, hormone receptor–positive metastatic breast cancers when combined with antiestrogen therapy. To uncover mechanisms of resistance, we conducted a detailed, longitudinal analysis of tumor and plasma circulating tumor DNA (ctDNA) among such patients from a phase I/II trial combining alpelisib with an aromatase inhibitor (NCT01870505). The trial’s primary objective was to establish safety with maculopapular rash emerging as the most common grade 3 adverse event (33%). Among 44 evaluable patients, the observed clinical benefit rate was 52%. Correlating genetic alterations with outcome, we identified loss-of-function PTEN mutations in 25% of patients with resistance. ESR1 activating mutations also expanded in number and allele fraction during treatment and were associated with resistance. These data indicate that genomic alterations that mediate resistance to alpelisib or antiestrogen may promote disease progression and highlight PTEN loss as a recurrent mechanism of resistance to PI3Kα inhibition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The assembled prospective somatic mutational data from ctDNA and tumors for the entire cohort have been deposited for visualization and download in the cBioPortal for Cancer Genomics (http://cbioportal.org/) under the following URL: https://www.cbioportal.org/study/summary?id=breast_alpelisib_2020.
Source data for Figs. 1–5 and for Extended Data Figs. 1 and 3–5 are available online.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The custom codes written to analyze the cfDNA data are available at https://github.com/ndbrown6/MSK-BYL-NC
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Ma, C. X., Reinert, T., Chmielewska, I. & Ellis, M. J. Mechanisms of aromatase inhibitor resistance. Nat. Rev. Cancer 15, 261–275 (2015).
Toy, W. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Schiavon, G. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 7, 313ra182 (2015).
Razavi, P. et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438 (2018).
Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Isakoff, S. J. et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 65, 10992–11000 (2005).
Lauring, J., Park, B. H. & Wolff, A. C. The phosphoinositide-3-kinase-Akt-mTOR pathway as a therapeutic target in breast cancer. J. Natl Compr. Canc. Netw. 11, 670–678 (2013).
Carlo, M. I. et al. A phase Ib study of BEZ235, a dual inhibitor of phosphatidylinositol 3-kinase (PI3K) and mammalian target of rapamycin (mTOR), in patients with advanced renal cell carcinoma. Oncologist 21, 787–788 (2016).
Sarker, D. et al. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 21, 77–86 (2015).
Juric, D. et al. Phase I dose-escalation study of taselisib, an oral PI3K inhibitor, in patients with advanced solid tumors. Cancer Discov. 7, 704–715 (2017).
Bohnacker, T. et al. Deconvolution of buparlisib’s mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention. Nat. Commun. 8, 14683 (2017).
Bendell, J. C. et al. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 30, 282–290 (2012).
Baselga, J. et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 904–916 (2017).
Bosch, A. et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci. Transl. Med. 7, 283ra251 (2015).
Fox, E. M., Kuba, M. G., Miller, T. W., Davies, B. R. & Arteaga, C. L. Autocrine IGF-I/insulin receptor axis compensates for inhibition of AKT in ER-positive breast cancer cells with resistance to estrogen deprivation. Breast Cancer Res. 15, R55 (2013).
Toska, E. et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 355, 1324–1330 (2017).
Creighton, C. J. et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 12, R40 (2010).
Juric, D. et al. Phosphatidylinositol 3-kinase α-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: results from the first-in-human study. J. Clin. Oncol. 36, 1291–1299 (2018).
Juric, D. et al. Alpelisib plus fulvestrant in PIK3CA-altered and PIK3CA-wild-type estrogen receptor-positive advanced breast cancer: a phase 1b clinical trial. JAMA Oncol. 5, e184475 (2018).
Mayer, I. A. et al. A phase Ib study of alpelisib (BYL719), a PI3Kα-specific inhibitor, with letrozole in ER+/HER2-negative metastatic breast cancer. Clin. Cancer Res. 23, 26–34 (2017).
Andre, F. et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
Will, M. et al. Rapid induction of apoptosis by PI3K inhibitors is dependent upon their transient inhibition of RAS-ERK signaling. Cancer Discov. 4, 334–347 (2014).
Mayer, I. A. et al. A phase II randomized study of neoadjuvant letrozole plus alpelisib for hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer (NEO-ORB). Clin. Cancer Res. 25, 2975–2987 (2019).
Baselga, J. et al. Phase III study of taselisib (GDC-0032)+ fulvestrant (FULV) v FULV in patients (pts) with estrogen receptor (ER)-positive, PIK3CA-mutant (MUT), locally advanced or metastatic breast cancer (MBC): primary analysis from SANDPIPER. J. Clin. Oncol. 36, LBA1006 (2018).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. https://doi.org/10.1200/PO.17.00011 (2017).
Juric, D. et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518, 240–244 (2015).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Ng, C. K. Y. et al. Genetic heterogeneity in therapy-naive synchronous primary breast cancers and their metastases. Clin. Cancer Res. 23, 4402–4415 (2017).
Murtaza, M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
De Mattos-Arruda, L. et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann. Oncol. 25, 1729–1735 (2014).
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
Odegaard, J. I. et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin. Cancer Res. 24, 3539–3549 (2018).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 10, e0140712 (2015).
Rodriguez-Escudero, I. et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum. Mol. Genet. 20, 4132–4142 (2011).
Marsh, D. J. et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan–Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum. Mol. Genet. 7, 507–515 (1998).
Han, S. Y. et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 60, 3147–3151 (2000).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Castel, P. & Scaltriti, M. in Resistance to Anti-Cancer Therapeutics Targeting Receptor Tyrosine Kinases and Downstream Pathways (eds. Yarden, Y. & Elkabets, M.) 117–146 (Springer International Publishing, 2018).
Zill, O. A. et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 24, 3528–3538 (2018).
Siravegna, G. et al. Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell 34, 148–162 e147 (2018).
Jia, S. et al. Essential roles of PI(3)K-p110β in cell growth, metabolism and tumorigenesis. Nature 454, 776–779 (2008).
Wee, S. et al. PTEN-deficient cancers depend on PIK3CB. Proc. Natl Acad. Sci. USA 105, 13057–13062 (2008).
Spoerke, J. M. et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 7, 11579 (2016).
Chandarlapaty, S. et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2, 1310–1315 (2016).
Ng, C. K. et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 16, 107 (2015).
Toledo, R. A. et al. Exome sequencing of plasma DNA portrays the mutation landscape of colorectal cancer and discovers mutated VEGFR2 receptors as modulators of antiangiogenic therapies. Clin. Cancer Res. 24, 3550–3559 (2018).
Chicard, M. et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin. Cancer Res. 24, 939–949 (2018).
Frenel, J. S. et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin. Cancer Res. 21, 4586–4596 (2015).
Keup, C. et al. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell. Mol. Life Sci. 77, 497–509 (2019).
Cheng, D. T. et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Cheng, D. T. et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med. Genomics 10, 33 (2017).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Acknowledgements
This work was supported by Novartis Pharmaceuticals and National Institutes of Health awards P30 CA008748, R01 CA190642 (M.S.), R01 CA234361 (D.B.S. and S.C.), R01CA204999 (S.C.), the Breast Cancer Alliance Young Investigator Award (P.R.), Conquer Cancer Foundation Young Investigator Award (P.D.S.), the Breast Cancer Research Foundation (M.S., J.S.R.-F. and S.C.), Damon Runyon Cancer Research Foundation (S.C.), Stand Up to Cancer (M.S., Cancer Drug Combination Convergence Team), the V Foundation (M.S.), the National Science Foundation (M.S.), the Geoffrey Beene Cancer Research Center and a kind gift from B. Smith.
Author information
Authors and Affiliations
Contributions
Authors contributed as follows: study conception: P.R., M.N.D., P.D.S., M.E.M., M.S. and S.C.; data acquisition: P.R., M.N.D., P.D.S., W.T., B.T.L., N.V., S.M., K.J., B.A.C., P.S., S.Z., A.C., E.C., S.M.S., A.C., M.F.B., R.J.N., J.I.O., R.B.L., D.B.S., M.E.L., E.B. and M.E.M.; data analysis and interpretation: P.R., D.N.B., H.H.W., R.S., S.P., A.S., R.J.N., J.I.O. and J.S.R.-F.; bioinformatics and genomic analysis: P.R., D.N.B., H.H.W., R.S., P.S., A.S. and J.S.R.-F.; manuscript first draft: P.R., M.N.D., P.D.S. and M.E.M. S.C. wrote the manuscript with input from all authors. All authors were responsible for manuscript review and approval.
Corresponding authors
Ethics declarations
Competing interests
P.R. reports consulting or advisory role for Novartis, AstraZeneca and Foundation Medicine and institutional research support from Illumina and GRAIL; M.N.D. is an employee of Eli Lilly; P.D.S. reports consulting with Tmunity and research funding from AstraZeneca; B.T.L. reports consulting/advisory board for Genentech, ThermoFisher Scientific, Guardant Health, Hengrui Therapeutics, Mersana Therapeutics, Biosceptre Australia and institutional research support from Illumina, GRAIL, Genentech and AstraZeneca; N.V. reports a consulting or advisory role for Novartis; K.J. reports a consulting or advisory role for ADC Therapeutics, AstraZeneca, Jounce Therapeutics, Novartis, Pfizer, Spectrum and Taiho and research funding from ADC Therapeutics (Inst), Clovis Oncology (Inst), Debio (Inst), Genentech (Inst), Novartis (Inst), Novita (Inst), Pfizer (Inst) and other relationships with Jounce Therapeutics, Novartis, Pfizer and Taiho; A.C. reports being an advisory board member for Accurate Medical and a Stockholder for Amgen; L.N. reports honoraria from Advanced Breast Cancer 4 International Consensus Conference, Bluprint Oncology Concepts, Celgene, Context Therapeutics and MCI Breast Cancer Symposium, consulting or advisory roles for Advanced Breast Cancer International Consensus Conference, Bluprint Oncology Concepts, Celgene, Context Therapeutics and MCI Breast Cancer Symposium and travel and accommodations expenses from Advanced Breast Cancer International Consensus Conference, Celgene and MCI Breast Cancer Symposium; A.S., R.J.N., J.I.O. and R.B.L. are employees and stockholders of Guardant Health; M.E.R. reports honoraria from AstraZeneca, consulting or advisory roles for AstraZeneca, McKesson, Merck and Pfizer, research funding from Abbvie (Inst), AstraZeneca (Inst), InVitae (Inst), Medivation (Inst), Myriad Genetics (Inst) and Tesaro (Inst) and travel and accommodations expenses from AstraZeneca; M.E.L. reports serving as a consultant or speaking for Legacy Healthcare Services, Adgero Bio, Amryt, Celldex, Debiopharm, Galderma, Johnson & Johnson, Novocure, Lindi, Merck, Sharp and Dohme Corp, Helsinn Healthcare SA, Janssen Research & Development LLC, Menlo Therapeutics, Novartis, Roche, AbbVie, Boehringer Ingelheim, Allergan, Amgen, E. R. Squibb & Sons, LLC, EMD Serono, AstraZeneca, Genentech, Leo Pharma, Seattle Genetics, Bayer, Manner, SAS, Lutris, Pierre Fabre, Paxman Coolers, Adjucare, Dignitana, Biotechspert, Teva Mexico, Parexel, OnQuality, Novartis, Our Brain Bank and Takeda Millenium; and receiving research funding from Veloce, US Biotest, Berg, BMS, Lutris, Paxman and Novocure; J.S.R.-F. reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Roche, Genentech and Invicro; D.B.S. received honoraria and consulted for Pfizer, Loxo, Vivideon, Illumina and Lilly Oncology; M.S. is on the Advisory Board of the Bioscience Institute and Menarini Ricerche, received research funds from Puma Biotechnology, Daiichi Sankyo, Targimmune, Immunomedics and Menarini Ricerche, is a co-founder of Medendi Medical Travel and received honoraria from Menarini Ricerche and ADC; S.C. reports institutional research funding from Novartis, Eli Lilly, Sanofi, Daiichi Sankyo and Genentech and ad hoc consulting for Novartis, Context Therapeutics, Sermonix, Eli Lilly, BMS and Revolution Medicine.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
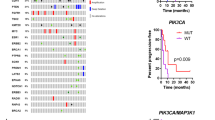
Extended Data Fig. 1 Characterization of ctDNA variants.
a, Comparison of variant allele fraction (VAF) of PIK3CA mutations measured using the targeted ctDNA assay (y-axis) and ddPCR (x-axis). cfDNA samples extracted from 65 samples (35 patients) with canonical hotspot PIK3CA mutations were subjected to ddPCR. An aliquot of the same cfDNA isolate was used for targeted DNA assay (G360 assay, Guardant Health, CA). b, ctDNA VAFs of all the somatic variants detected in ctDNA restricted to 38 patients with available tumor next generation sequencing results. Colors indicate the altered gene and black borders indicate whether the alteration was detected in the tumor tissue. c, Comparison of VAF of mutations detected in the pre-treatment and post-treatment ctDNA samples of six patients with evaluable paired ctDNA specimens and PTEN loss-of-function mutations in either sample. The colors of the circles indicate mutated gene.
Extended Data Fig. 2 Normalized ∆VAF and ∆CCF model.
Toy model showing the calculation of a, normalized ∆VAF and b, ∆CCF for a fictitious pair of pre- and post-treatment ctDNA samples. Three mutations are shown; PIK3CA, ESR1 and NF1. The canonical PIK3CA mutation is expected to be clonal in the pre- and post-treatment ctDNA samples. The difference in VAF of PIK3CA between the pair is a surrogate of difference in purity and tumor burden. To quantify the change in VAF or CCF of other mutations in addition to what is expected from the difference in purity or tumor burden, we calculate the Log10 difference between the post-treatment cfDNA and the value obtained from the regression that is in the examples above, observed post-treatment NF1 VAF – expected post-treatment NF1 VAF. In both cases, the regression has zero y-intercept.
Extended Data Fig. 3 Change in variant allele fraction (∆VAF) of all mutations in post- vs pre-treatment ctDNA specimens.
Heatmap of change in VAF comparing the post-treatment with pre-treatment ctDNA of 32 evaluable patients (n=64 samples) normalized according to the change in ctDNA fraction as a proxy of change in disease burden. The size of the boxes represents the relative change and the color gradient of the boxes represent increase or decrease in ∆VAF. The top section shows the time to treatment failure (weeks), reason off study, and the best response on therapy. Multiple mutations in a same gene are indicated, for example patient #47 had two different PIK3CA mutations with one mutation having no change while the other one expanded in the post-treatment sample (positive ∆VAF). PR: partial response, SD: stable disease, PD: progressive disease, NE: not evaluable for response.
Extended Data Fig. 4 Comparison of normalized change in ctDNA mutations.
a, Comparison of normalized change in ctDNA mutations (∆VAF) with relative change in mutations cancer cell fractions (∆CCF). The analysis includes 256 mutations. b, Comparison of estimated CCF based on combined tumor and ctDNA approach (Methods) with CCF estimated based on ctDNA-only approach54,55. The analysis includes 379 mutations. Pearson’s correlation coefficients (R) and two-sided p values are provided.
Extended Data Fig. 5 Mutational signatures in two hypermutated cases with acquired PTEN mutations.
96 base substitution profiles of pre-treatment ctDNA samples from the two hypermutated cases that eventually developed PTEN mutations under therapy showing dominant APOBEC signatures (Signatures 2 and 13).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Supplementary Table 3
Sequences of oligonucleotides used for making Y537S knock in MCF7 cell line
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Razavi, P., Dickler, M.N., Shah, P.D. et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat Cancer 1, 382–393 (2020). https://doi.org/10.1038/s43018-020-0047-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-0047-1
This article is cited by
-
p4EBP1 staining predicts outcome in ER-positive endocrine-resistant metastatic breast cancer patients treated with everolimus and exemestane
British Journal of Cancer (2024)
-
Comparative genomic analysis of PIK3R1-mutated and wild-type breast cancers
Breast Cancer Research and Treatment (2024)
-
Nuclear lncRNA NORSF reduces E2 release in granulosa cells by sponging the endogenous small activating RNA miR-339
BMC Biology (2023)
-
The emerging role of PI3K inhibitors for solid tumour treatment and beyond
British Journal of Cancer (2023)
-
Source, co-occurrence, and prognostic value of PTEN mutations or loss in colorectal cancer
npj Genomic Medicine (2023)