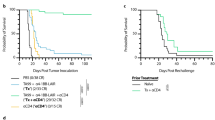
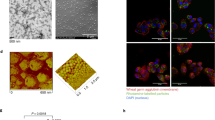
Abstract
Innate pattern recognition receptor agonists, including Toll-like receptors (TLRs), alter the tumor microenvironment and prime adaptive antitumor immunity. However, TLR agonists present toxicities associated with widespread immune activation after systemic administration. To design a TLR-based therapeutic suitable for systemic delivery and capable of safely eliciting tumor-targeted responses, we developed immune-stimulating antibody conjugates (ISACs) comprising a TLR7/8 dual agonist conjugated to tumor-targeting antibodies. Systemically administered human epidermal growth factor receptor 2 (HER2)-targeted ISACs were well tolerated and triggered a localized immune response in the tumor microenvironment that resulted in tumor clearance and immunological memory. Mechanistically, ISACs required tumor antigen recognition, Fcγ-receptor-dependent phagocytosis and TLR-mediated activation to drive tumor killing by myeloid cells and subsequent T-cell-mediated antitumor immunity. ISAC-mediated immunological memory was not limited to the HER2 ISAC target antigen since ISAC-treated mice were protected from rechallenge with the HER2− parental tumor. These results provide a strong rationale for the clinical development of ISACs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The NanoString gene expression data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession no. GSE157870. The source data for the structures shown in Extended Data Fig. 1c is available on the Protein Data Bank with accession no. 3W3N. All other source data of the figures and extended figures, as well as supporting experiments, are available from the corresponding author upon reasonable request. Biological materials will be made available upon reasonable request. Source data are provided with this paper.
Code availability
Code for the DREMI algorithm is available at https://github.com/KrishnaswamyLab/scprep/. The R code for the CyTOF analysis and to generate volcano plots utilizing nSolver-derived P values is available upon reasonable request.
References
Davis, T. A. et al. Rituximab anti-CD20 monoclonal antibody therapy in non-Hodgkin’s lymphoma: safety and efficacy of re-treatment. J. Clin. Oncol. 18, 3135–3143 (2000).
Grillo-López, A. J. et al. Rituximab: the first monoclonal antibody approved for the treatment of lymphoma. Curr. Pharm. Biotechnol. 1, 1–9 (2000).
Fridman, W. H., Pagès, F., Sautès-Fridman, C. & Galon, J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012).
Galon, J. & Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18, 197–218 (2019).
Gabrilovich, D. I., Ostrand-Rosenberg, S. & Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 12, 253–268 (2012).
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015).
Urban-Wojciuk, Z. et al. The role of TLRs in anti-cancer immunity and tumor rejection. Front. Immunol. 10, 2388 (2019).
Carmi, Y. et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature 521, 99–104 (2015).
Carmi, Y. et al. Akt and SHP-1 are DC-intrinsic checkpoints for tumor immunity. JCI Insight 1, e89020 (2016).
Sagiv-Barfi, I. et al. Eradication of spontaneous malignancy by local immunotherapy. Sci. Transl. Med. 10, eaan4488 (2018).
Spitzer, M. H. et al. Systemic immunity is required for effective cancer immunotherapy. Cell 168, 487–502.e15 (2017).
Jurk, M. et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 3, 499 (2002).
Saitoh, S.-I. et al. TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat. Commun. 8, 1592 (2017).
Mattson, G. et al. A practical approach to crosslinking. Mol. Biol. Rep. 17, 167–183 (1993).
Gabay, C., Ben-Bassat, H., Schlesinger, M. & Laskov, R. Somatic mutations and intraclonal variations in the rearranged Vk genes of B-non-Hodgkin’s lymphoma cell lines. Eur. J. Haematol. 63, 180–191 (1999).
Alonso, M. N. et al. TH1, TH2, and TH17 cells instruct monocytes to differentiate into specialized dendritic cell subsets. Blood 118, 3311–3320 (2011).
Clarke, S. R. et al. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol. Cell Biol. 78, 110–117 (2000).
Zehn, D., Lee, S. Y. & Bevan, M. J. Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214 (2009).
Kondratova, M. et al. A multiscale signalling network map of innate immune response in cancer reveals cell heterogeneity signatures. Nat. Commun. 10, 4808 (2019).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Kawai, T. & Akira, S. TLR signaling. Cell Death Differ. 13, 816–825 (2006).
Sánchez-Mejorada, G. & Rosales, C. Signal transduction by immunoglobulin Fc receptors. J. Leukoc. Biol. 63, 521–533 (1998).
Krishnaswamy, S. et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014).
Kiefer, F. et al. The Syk protein tyrosine kinase is essential for Fcγ receptor signaling in macrophages and neutrophils. Mol. Cell. Biol. 18, 4209–4220 (1998).
Braselmann, S. et al. R406, an orally available spleen tyrosine kinase inhibitor blocks Fc receptor signaling and reduces immune complex-mediated inflammation. J. Pharmacol. Exp. Ther. 319, 998–1008 (2006).
Bruhns, P. et al. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009).
Nimmerjahn, F. & Ravetch, J. V. Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 310, 1510–1512 (2005).
Jefferis, R. Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends Pharmacol. Sci. 30, 356–362 (2009).
Li, J. Y. et al. A biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell 29, 117–129 (2016).
Luque-Cabal, M., García-Teijido, P., Fernández-Pérez, Y., Sánchez-Lorenzo, L. & Palacio-Vázquez, I. Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin. Med. Insights Oncol. 10, 21–30 (2016).
Tanner, M. et al. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol. Cancer Ther. 3, 1585–1592 (2004).
Xia, Z. et al. Innate immune response to human bone marrow fibroblastic cell implantation in CB17 scid/beige mice. J. Cell. Biochem. 98, 966–980 (2006).
Lewis Phillips, G. D. et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 68, 9280–9290 (2008).
Ning, S., Pagano, J. S. & Barber, G. N. IRF7: activation, regulation, modification and function. Genes Immun. 12, 399–414 (2011).
Cao, Q. et al. Renal F4/80+ CD11c+ mononuclear phagocytes display phenotypic and functional characteristics of macrophages in health and in adriamycin nephropathy. J. Am. Soc. Nephrol. 26, 349–363 (2015).
Sheng, J. et al. A discrete subset of monocyte-derived cells among typical conventional type 2 dendritic cells can efficiently cross-present. Cell Rep. 21, 1203–1214 (2017).
Moynihan, K. D. et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat. Med. 22, 1402–1410 (2016).
Muller, W. J., Sinn, E., Pattengale, P. K., Wallace, R. & Leder, P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105–115 (1988).
Lee, J. et al. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl Acad. Sci. USA 103, 1828–1833 (2006).
Bonaventura, P. et al. Cold tumors: a therapeutic challenge for immunotherapy. Front. Immunol. 10, 168 (2019).
Guilliams, M., Bruhns, P., Saeys, Y., Hammad, H. & Lambrecht, B. N. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 14, 94–108 (2014).
Nimmerjahn, F. & Ravetch, J. V. Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 (2008).
Regnault, A. et al. Fcγ receptor-mediated induction of dendritic cell maturation and major histocompatibility complex class I-restricted antigen presentation after immune complex internalization. J. Exp. Med. 189, 371–380 (1999).
Rafiq, K., Bergtold, A. & Clynes, R. Immune complex-mediated antigen presentation induces tumor immunity. J. Clin. Invest. 110, 71–79 (2002).
Genentech. Herceptin (trastuzumab). Package Insert (Roche, 2018).
Safina, B, & Lee, A. Protoc. Exch. https://doi.org/10.21203/rs.3.pex-1149/v1 (2020)..
Zunder, E. R. et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015).
Acknowledgements
We thank P. Anand, S. Deol and M. Zhou for their bioconjugation expertise, L. B. Torrez Dulgeroff for experimental expertise and P. Basto, N. E. Reticker-Flynn, T. Prestwood and B. Mallet for helpful discussions. We also thank the Stanford Blood Center, specifically L. Tolentino, O. Choi, N. Wu and K. Kealoha, for their support and experimental expertise. We extend our appreciation to M. L. Disis and D. Cecil from the University of Washington who provided the MMC tumor cell line. We extend our warmest thanks to the Stanford blood donors who save lives and made this work possible. Funding was provided by a Stanford BioX Bowes Fellowship (S.E.A.), EMBO Long-Term Fellowship no. ALTF 1141-2017 (F.J.H.), Swiss National Science Foundation Early Postdoc Mobility Fellowship no. P2ZHP3-171741 (F.J.H.), National Institutes of Health (NIH)/National Institute and General Medical Sciences Cell and Molecular Biology Training grant no. T32GM007276 (S.C.K.) and NIH grant nos. CA209971 (E.G.E.), CA222969 (E.G.E.) and 1DP2OD022550-01 (S.C.B.).
Author information
Authors and Affiliations
Contributions
S.E.A., E.G.E. and M.N.A. conceptualized the study. S.E.A., C.I.P., J.D.G., J.C.G., F.J.H., J.A.K., S.C.K., J.C.P., B.A., V.C.R., S.J.C., S.C.B., D.D. and M.N.A. devised the experimental methodology. S.E.A., C.I.P., J.D.G., J.C.G., J.A.K., F.J.H., Angela Luo, P.Y.H., L.K.B., S.C.K., Andrew Luo, M.L.N., L.Y.S., A. Lee, H. Lai, J.M., R.P.L., V.C.R., K.A.H., D.Y.J., S.J.C. and M.N.A. carried out the investigation. S.E.A., C.I.P., J.D.G., J.C.G., H. LeBlanc, L.K.B., D.D., E.G.E. and M.N.A. wrote the manuscript. B.S.S., M.K., G.Y., B.H.D., Y.C., S.C.B., D.D., E.G.E. and M.N.A. supervised the study.
Corresponding author
Ethics declarations
Competing interests
All authors affiliated with Bolt Biotherapeutics are current or former employees. Bolt Biotherapeutics-affiliated authors and Y.C. may hold stock options and/or equity in Bolt Biotherapeutics. E.G.E. is a founder and shareholder of Bolt Biotherapeutics and a member of the company’s board of directors. S.E.A., J.A.K., A. Lee, D.Y.J., E.G.E. and M.N.A. are Stanford- and/or Bolt Biotherapeutics-affiliated inventors of PCT/US2017/041268, which discloses T785-containing ISACs.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 T785 synthesis, design and activity on pDCs.
a, b, Confirmation of T785 structure via (a) proton NMR and (b) LC-MS. c, Molecular docking studies using a co-crystal structure of human TLR8 and R848 (PDB ID: 3W3N) to visualize T785 in the binding pocket of TLR8 supported solvent accessibility of the point of conjugation. Zoomed in views of the binding pocket show solvent accessibility of the exit vector (upper panel) and a view of the entire TLR8 protein with a single T785 docked (lower panel). d, HEK293 Null reporter cells, the parental cell line used for hTLR7 and hTLR8 reporter cells, were stimulated overnight with a concentration titration of T785 or R848 and activity was measured using QuantiBlue detection medium. Data are shown from one independent experiment. e, pDCs were isolated from healthy donor blood and stimulated overnight with T785 (TLR7/8), R848 (TLR7/8) or CpG (TLR9). IFNα secretion was measured by ELISA. Data shown (mean and SEM) are from n=3 independent donors.
Extended Data Fig. 2 Myeloid cell isolation and rituximab T785-ISAC activation.
a, Purity of freshly isolated myeloid APCs following negative selection by density gradient centrifugation using the RosetteSep Human Monocyte Enrichment Kit followed by magnetic separation with the EasySep Human Monocyte Enrichment Kit without CD16 Depletion as assessed by flow cytometry. b, Gating strategy for analysis of activation and differentiation of myeloid APCs in all described in vitro experiments. First, cells are gated as singlets by FSC or SSC to remove doublets. Next, a cellular size gate using SSC and FSC removes debris. Viable cells are then gated based on a Live/Dead stain. Finally, myeloid APCs are gated based on CD11c+ and HLA-DR+ expression. c, Histogram analysis of cell surface marker expression of total myeloid APCs following stimulation with 80 nM rituximab-ISAC or 80 nM rituximab. Data shown are from one independent donor and are representative of 3 independent experiments. d, The frequency of positive subsets (percentage positive) was calculated based on gating of unstimulated myeloid APCs in the absence of tumor. Panels above show the gating strategy for unstimulated myeloid APCs co-cultured with Toledo CD20+ tumor cells. A dose dependent response analogous to that measured with MFI (as reported in Fig. 1 and elsewhere) is observed when computed as percent positive of total myeloid APCs. Data shown (mean and SEM) are from n=3 independent donors and are representative of 3 independent experiments.
Extended Data Fig. 3 Myeloid APC differentiation and activation, antigen cross-presentation and T cell proliferation following ISAC stimulation.
a-c, Freshly isolated human myeloid APCs were cultured with rituximab, T785, rituximab and T785 or the rituximab T785-ISAC in the presence of CFSE-labeled CD20+ Toledo tumor cells at a 3:1 ratio. The rituximab concentration is depicted on the X-axis; T785 concentration is equivalent to the amount of T785 conjugated to the rituximab T785-ISAC. Myeloid APCs were analyzed after 18 hours via (a, b) flow cytometry or (c) cytokine bead array. d, Frozen myeloid APCs were thawed, rested for two hours and incubated with trastuzumab or trastuzumab T785-ISAC for 18 hours prior to assessment of cell surface markers by flow cytometry. a-d, Data are shown (mean and SEM) with 3 independent donors and are representative of 3 experiments; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 calculated by two-way ANOVA with Tukey’s multiple comparison correction. e, Mouse splenocytes were isolated and cultured in vitro with ovalbumin antigen complexed with anti-OVA CL264-ISAC or anti-OVA mAb. Antigen cross-presentation was measured using multicolor flow cytometry through detection of MHC-I bound SIINFEKL peptide on total CD11c+ splenic cells. CD8+ T cells with OVA-specific TCR were isolated from OT-1 transgenic mice and co-cultured with anti-OVA mAb or anti-OVA ISAC stimulated APCs. T cell proliferation was measured by final T cell count with the initial number of APCs subtracted.
Extended Data Fig. 4 ISACs elicit distinct intracellular signaling in monocytes and cDCs.
a-c,e-f, Freshly isolated human PBMC were stimulated with 1 μM of rituximab-ISAC or an equimolar mixture of rituximab and T785 in the presence of CD20+ Toledo tumor cells at a 1:1 ratio for 15 minutes. a, Signaling induction of p-MAPKAPK2, p-p38, p-CREB and p-S6 in monocytes and cDCs was quantified by the arcsinh of the ISAC or mixture as compared to the unstimulated PBMCs (n=6 independent donors). b, cDCs were subsetted as CD141+ cDC1s and CD1c+ cDC2s (n=6 independent donors). c, Signaling through pERK1/2 and p-S6 measured following 5, 15, or 30-minute stimulation with rituximab T785-ISAC or the mixture of rituximab and T785 (n=2 independent donors). d, Monocytes were treated with 1 μM of R406 prior to stimulation with 2 μM T785 overnight. Activation was assessed by flow cytometry, and no statistical significance was seen between the conditions with and without Syk inhibition. The dashed line represents the expression level of the unstimulated control (n=2 technical replicates of pooled myeloid APCs with 6 independent donors per pool). e, Signaling induction of p-MAPKAPK2, pERK1/2 and pIRF7 was quantified by the arcsinh of the ISAC with and without Syk blockade (R406) as compared to the unstimulated control (n=6 independent donors). f, Signaling induction of pIRF7 was quantified by the arcsinh following stimulation with rituximab T785-ISAC or rituximab-TLRnull-ISAC (TLRnull-ISAC) (n=6 independent donors). a, b, d-f, Data shown (mean and SEM) are representative of two independent experiments. P values were calculated by two-tailed paired T tests.
Extended Data Fig. 5 Analytical characterization of ISACs and assessment of inhibition of in vitro cancer cell proliferation.
a, LC/MS characterization of trastuzumab T785-ISAC and rituximab T785-ISAC. b, Analytical SEC and endotoxin measurement of trastuzumab T785-ISAC and rituximab T785-ISAC. c, LC/MS analysis of trastuzumab T785-ISAC compared to the trastuzumab CL264-ISAC. Conjugates were also found to be endotoxin low, with less than 0.05 EU/mg detected. d, Cancer cell line proliferation assayed with an MTT assay 72 hours after incubation with the indicated tumor cell line and the indicated test article. Rituximab-ISACs were utilized as isotype controls. Data are from 2 independent experiments performed in triplicate.
Extended Data Fig. 6 T785-ISACs are efficacious in the HCC1954 and JIMT-1 tumor models.
a, In vitro HER2 expression of the HCC1954 tumor cell line assessed by flow cytometry with PE-labeled anti-HER2 antibody (clone 24D2) (blue) or the isotype control (red). b-e, Spider plots visualizing tumor growth kinetics and survival for individual animals which are shown as mean tumor volume in Fig. 4. f, In vitro HER2 expression of the JIMT-1 tumor cell line assessed by flow cytometry with PE-labeled anti-HER2 antibody (clone 24D2) (blue) or the isotype control (red). g-h, Spider plots visualizing tumor growth kinetics and survival for individual animals which are shown as mean tumor volume in Fig. 4.
Extended Data Fig. 7 Gene expression data for genes used to calculate gene signatures in the HCC1954 model.
NanoString gene expression data was obtained using the mouse pan-cancer immune profiling panel, and gene expression pathway signature analysis was performed using the nSolver Advanced Analysis Pathway Score algorithm. Data are shown as heat maps with n=5 mice per group for all genes used to calculate gene signature scores for (a) Dendritic Cell Functions, Macrophage Functions, Antigen Processing and Presentation, (b) Chemokines and (c) Cytokines.
Extended Data Fig. 8 Characterization of the MMC syngeneic tumor cell line model.
a, rHER2 expression measured on the MMC tumor cell line by flow cytometry with fluorescently conjugated anti-rHER2 antibody (red) or an isotype control (blue) grown in vitro (left) or assessed 23 days post implantation in FVB/N-TgN (MMTV-Erbb2) mice (right). b, Body weight (left; mean and SEM) and tumor volume (right) following treatment with 5 mg/kg of the indicated test article (n=5 mice per group) measured at time of harvest for analysis by NanoString, MSD protein quantification, or immunohistochemistry. c, Kaplan-Meier curves representing the data shown in Fig. 6a–c. d, Mice were pre-treated with clodronate-loaded or control liposomes for phagocyte depletion then treated IP with test articles at 5 mg/kg q5d x 2 (n=5 mice per group). Tumors were harvested at day 6 and were analyzed by flow cytometry, with myeloid APCs defined as viable cells with CD45+CD11b+Ly6G−CD11c+F4/80+ expression. b, d, For box-and-whisker plots, boxes represent 25th and 75th percentiles, center lines indicate median values and whiskers represent minimum and maximum values. Statistical significance was calculated by (b) two-way or (d) one-way ANOVA with Tukey’s multiple comparison correction. e, Gating scheme for flow cytometry analysis of tumor samples from MMC syngeneic tumor model.
Extended Data Fig. 9 Gene expression data for genes used to calculate gene signatures in the MMC model.
NanoString gene expression data were obtained using the mouse pan-cancer immune profiling panel, and gene expression pathway signature analysis was performed using the nSolver Advanced Analysis Pathway Score algorithm. Data are shown as heat maps (n=5 mice per group) for genes used to calculate gene signature scores for (a) Dendritic Cell Functions, Macrophage Functions, Antigen Processing and Presentation, (b) Chemokines, (c) Cytokines, and (d) T Cell Functions.
Extended Data Fig. 10 Growth kinetics, tolerability and myeloid activation in mouse tumor models treated with ISACs.
a-b, Spider plots visualizing the tumor growth kinetics for individual animals in HCC1954 and JIMT-1 xenograft tumor studies comparing trastuzumab T785-ISAC and trastuzumab CL264-ISAC (shown in Fig. 8c, d). c, SCID/beige mice were implanted with HCC1954 tumor cells and randomized when the tumor volume reached 50–75 mm3 (n=5 mice per group). Rag2/IL2rg knockout mice were implanted with JIMT-1 tumor cells and randomized when the tumor volume reached 75–150 mm3 (n=3 mice per group). Mice were treated IP with 5 mg/kg of trastuzumab, trastuzumab T785-ISAC, trastuzumab CL264-ISAC or the respective isotype ISACs, and serum cytokine levels were measured by ELISA 4 hours following administration. Body weight was measured following dosing every 5 days, with doses indicated by dashed vertical lines. d, FVB/N-TgN (MMTV-Erbb2) mice were implanted with the MMC tumor cell line. When tumors reached an average volume of (left) 500 mm3 or (right) 300 mm3, mice were treated IP with 5 mg/kg of rHER2 mAb or rHER2 CL264-ISAC q5d x 3 (rHER2 mAb, n=3; rHER2 CL264-ISAC, n=4). For CD8 T cell depletion, mice were treated with an antibody to deplete CD8 T cells or with a rIgG2b control, and then treated IP with 2 mg/kg of rHER2 mAb or rHER2 CL264-ISAC q7d x 2 (n=7 mice per group, except rHER2 CL264-ISAC plus IgG2b isotype control (n=5)). e, rHER2 CL264-ISAC treated mice that experienced complete tumor regression for >90 days (n=4) were rechallenged with the MMC tumor cell line. Tumor naïve mice (n=5) were implanted with the MMC tumor line as controls. f, Body weight and survival of Balb/c mice implanted with the CT26-rHER2 tumor cell line and treated IP with 10 mg/kg of rHER2 mAb or rHER2 CL264-ISAC q5d x 6 (n=8). g, Kaplan-Meier curves demonstrating overall survival animals rechallenged with contralateral parental CT26 and 4T1 tumors. Survival across all groups was impacted by the growth of the genetically distinct 4T1 tumor. h, Gene expression of CXCL9 and CXCL11 in HCC1954 tumors following treatment with 5 mg/kg of trastuzumab, trastuzumab T785-ISAC, rituximab, or rituximab T785-ISAC 24 hours after treatment, as analyzed by Nanostring mRNA quantification (n=5 mice per group). i, Confirmation of cell depletion in the blood of HCC1954 tumor-bearing mice by flow cytometry 13 days following initiation of treatment. c-f, h, i, Data are shown as mean and SEM with statistical significance calculated by (d-e) two-way ANOVA with Tukey’s multiple comparison corrections, (h) one-way ANOVA with Tukey’s multiple comparison corrections, or (i) unpaired T tests with a correction for multiple-comparisons using the Holm-Sidak method; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Supplementary information
Supplementary Information
Supplementary Fig. 1
Supplementary Tables
Supplementary Tables 1 and 2
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Ackerman, S.E., Pearson, C.I., Gregorio, J.D. et al. Immune-stimulating antibody conjugates elicit robust myeloid activation and durable antitumor immunity. Nat Cancer 2, 18–33 (2021). https://doi.org/10.1038/s43018-020-00136-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-00136-x
This article is cited by
-
Drug conjugates for the treatment of lung cancer: from drug discovery to clinical practice
Experimental Hematology & Oncology (2024)
-
Exploring the next generation of antibody–drug conjugates
Nature Reviews Clinical Oncology (2024)
-
New immune cell engagers for cancer immunotherapy
Nature Reviews Immunology (2024)
-
The Streptococcus virulence protein PepO triggers anti-tumor immune responses by reprograming tumor-associated macrophages in a mouse triple negative breast cancer model
Cell & Bioscience (2023)
-
Payload diversification: a key step in the development of antibody–drug conjugates
Journal of Hematology & Oncology (2023)