Abstract
Metabolic reprogramming is a key hallmark of cancer, but less is known about metabolic plasticity of the same tumor at different sites. Here, we investigated the metabolic adaptation of leukemia in two different microenvironments, the bone marrow and the central nervous system (CNS). We identified a metabolic signature of fatty acid synthesis in CNS leukemia, highlighting stearoyl-CoA desaturase (SCD) as a key player. In vivo SCD overexpression increases CNS disease, whereas genetic or pharmacological inhibition of SCD decreases CNS load. Overall, we demonstrated that leukemic cells dynamically rewire metabolic pathways to suit local conditions and that targeting these adaptations can be exploited therapeutically.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data supporting this study’s findings have been deposited in GEO (accession number: GSE135115). The GSE135115 SuperSeries is entitled “Gene expression profiles of MLL-AF4 and TEL-AML1 acute lymphoblastic leukemia blasts retrieved from central nervous system and spleen”. This SuperSeries contains two series related to SEM and REH experiments as follows: GSE135113 “Gene expression profiles of MLL-AF4 acute lymphoblastic leukemia blasts retrieved from central nervous system and spleen” and GSE135114 “Gene expression profiles of TEL-AML1 acute lymphoblastic leukemia blasts retrieved from central nervous system and spleen”. Previously published human and primograft data re-analyzed here are available under accession codes GSE60926 and GSE89710. The source data associated with each figure are provided with the manuscript. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 (2011).
DeBerardinis, R. J. & Chandel, N. S. Fundamentals of cancer metabolism. Sci. Adv. 2, 1–18 (2016).
Cha, J.-Y. & Lee, H.-J. Targeting lipid metabolic reprogramming as anticancer therapeutics. J. Cancer Prev. 21, 209–215 (2017).
Gisselbrecht, C. Positron emission tomography – Guided therapy of aggressive non-Hodgkin lymphoma: Standard of care after the PETAL study? J. Clin. Oncol. 36, 3272–3273 (2018).
Caro, P. et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B-cell lymphoma. Cancer Cell 22, 547–560 (2012).
Kuntz, E. M. et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 23, 1234–1240 (2017).
Nachmias, B. & Schimmer, A. D. Metabolic flexibility in Leukemia—adapt or die. Cancer Cell 34, 695–696 (2018).
Olivares, O., Däbritz, J. H. M., King, A., Gottlieb, E. & Halsey, C. Research into cancer metabolomics: towards a clinical metamorphosis. Semin. Cell Dev. Biol. 43, 52–64 (2015).
Frishman-Levy, L. & Izraeli, S. Advances in understanding the pathogenesis of CNS acute lymphoblastic leukaemia and potential for therapy. Br. J. Haematol. 176, 157–167 (2017).
Pui, C. H. & Howard, S. C. Current management and challenges of malignant disease in the CNS in paediatric leukaemia. Lancet Oncol. 9, 257–268 (2008).
Halsey, C. et al. The impact of therapy for childhood acute lymphoblastic leukaemia on intelligence quotients; Results of the risk-stratified randomized central nervous system treatment trial MRC UKALL XI. J. Hematol. Oncol. 4, 1–12 (2011).
Iyer, N. S., Balsamo, L. M., Bracken, M. B. & Kadan-Lottick, N. S. Chemotherapy-only treatment effects on long-term neurocognitive functioning in childhood ALL survivors: a review and meta-analysis. Blood 126, 346–353 (2015).
Price, R. A. & Johnson, W. W. The central nervous system in childhood leukemia: I. The arachnoid. Cancer 31, 520–533 (1973).
Williams, M. T. S. et al. The ability to cross the blood-cerebrospinal fluid barrier is a generic property of acute lymphoblastic leukemia blasts. Blood 127, 1998–2006 (2016).
Bartram, J. et al. High throughput sequencing in acute lymphoblastic leukemia reveals clonal architecture of central nervous system and bone marrow compartments. Haematologica 103, e110–e114 (2018).
Kato, I. et al. Hypoxic adaptation of leukemic cells infiltrating the CNS affords a therapeutic strategy targeting VEGFA. Blood 129, 3126–3129 (2017).
Buonamici, S. et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 459, 1000–1004 (2009).
Krause, S. et al. Mer tyrosine kinase promotes the survival of t(1;19)-positive acute lymphoblastic leukemia (ALL) in the central nervous system (CNS). Blood 125, 820–830 (2015).
Cario, G. et al. High interleukin-15 expression characterizes childhood acute lymphoblastic leukemia with involvement of the CNS. J. Clin. Oncol. 25, 4813–4820 (2007).
Williams, M. T. S. et al. Interleukin-15 enhances cellular proliferation and upregulates CNS homing molecules in pre-B acute lymphoblastic leukemia. Blood 123, 3116–3127 (2014).
Frishman-Levy, L. et al. Central nervous system acute lymphoblastic leukemia: role of natural killer cells. Blood 125, 3420–3431 (2015).
Münch, V. et al. Central nervous system involvement in acute lymphoblastic leukemia is mediated by vascular endothelial growth factor. Blood 130, 643–654 (2017).
Spector, R., Robert Snodgrass, S. & Johanson, C. E. A balanced view of the cerebrospinal fluid composition and functions: focus on adult humans. Exp. Neurol. 273, 57–68 (2015).
Hühmer, A. F., Biringer, R. G., Amato, H., Fonteh, A. N. & Harrington, M. G. Protein analysis in human cerebrospinal fluid: physiological aspects, current progress and future challenges. Dis. Markers 22, 3–26 (2006).
Damkier, H. H., Brown, P. D. & Praetorius, J. Cerebrospinal fluid secretion by the choroid plexus. Physiol. Rev. 93, 1847–1892 (2013).
Méndez-Ferrer, S. et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 (2010).
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Olechnowicz, S. W. Z. & Edwards, C. M. Contributions of the host microenvironment to cancer-induced bone disease. Cancer Res. 74, 1625–1631 (2014).
Eckhoff, E. M., Queudeville, M., Debatin, K.-M. & Meyer, L. H. A novel B cell precursor ALL cell line (018Z) with prominent neurotropism and isolated CNS leukemia in a NOD/SCID/huALL xenotransplantation model. Blood 114, 1630–1630 (2009).
van der Velden, V. H. J. et al. New cellular markers at diagnosis are associated with isolated central nervous system relapse in paediatric B-cell precursor acute lymphoblastic leukaemia. Br. J. Haematol. 172, 769–781 (2016).
Theodoropoulos, P. C. et al. Discovery of tumor-specific irreversible inhibitors of stearoyl-CoA desaturase. Nat. Chem. Biol. 12, 218–225 (2016).
Metallo, C. M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2012).
Angelucci, C. et al. Pivotal role of human stearoyl-CoA desaturases (SCD1 and 5) in breast cancer progression: oleic acid-based effect of SCD1 on cell migration and a novel pro-cell survival role for SCD5. Oncotarget 9, 24364–24380 (2018).
Hess, D., Chisholm, J. W. & Igal, R. A. Inhibition of stearoyl-CoA desaturase activity blocks cell cycle progression and induces programmed cell death in lung cancer cells. PLoS ONE 5, e11394 (2010).
Wang, J. et al. High expression of stearoyl-CoA desaturase 1 predicts poor prognosis in patients with clear-cell renal cell carcinoma. PLoS ONE 11, e0166231 (2016).
Chen, L. et al. Stearoyl-CoA desaturase-1-mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci. Rep. 6, 1–11 (2016).
Kim, S. J., Choi, H., Park, S. S., Chang, C. & Kim, E. Stearoyl-CoA desaturase (SCD) facilitates proliferation of prostate cancer cells through enhancement of androgen receptor transactivation. Mol. Cells 31, 371–377 (2011).
Zhang, H., Li, H., Ho, N., Li, D. & Li, S. Scd1 plays a tumor-suppressive role in survival of leukemia stem cells and the development of chronic myeloid leukemia. Mol. Cell Biol. 32, 1776–1787 (2012).
Southam, A. D. et al. Drug redeployment to kill leukemia and lymphoma cells by disrupting SCD1-mediated synthesis of monounsaturated fatty acids. Cancer Res. 75, 2530–2540 (2015).
Imamura, K. et al. Discovery of novel and potent stearoyl coenzyme a desaturase 1 (SCD1) inhibitors as anticancer agents. Bioorganic Med. Chem. 25, 3768–3779 (2017).
Folger, O. et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7, 1–10 (2011).
Miyazaki, M., Man, W. C. & Ntambi, J. M. Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J. Nutr. 131, 2260–2268 (2001).
Brown, J. M. & Rudel, L. L. Stearoyl-coenzyme A desaturase 1 inhibition and the metabolic syndrome: considerations for future drug discovery. Curr. Opin. Lipidol. 21, 192–197 (2010).
Prieto, C. et al. NG2 antigen is involved in leukemia invasiveness and central nervous system infiltration in MLL-rearranged infant B-ALL. Leukemia 32, 633–644 (2018).
Pieters, R. et al. Outcome of infants younger than 1 year with acute lymphoblastic leukemia treated with the interfant-06 protocol: results from an international phase III randomized study. J. Clin. Oncol. 37, 2246–2256 (2019).
Vriens, K. et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 566, 403–406 (2019).
Ono, A. et al. Feedback activation of AMPK-mediated autophagy acceleration is a key resistance mechanism against SCD1 inhibitor-induced cell growth inhibition. PLoS ONE 12, e0181243 (2017).
Hagedorn, N. et al. Submicroscopic bone marrow involvement in isolated extramedullary relapses in childhood acute lymphoblastic leukemia: a more precise definition of “isolated” and its possible clinical implications, a collaborative study of the Resistant Disease Committee. Blood 110, 4022–4029 (2007).
Yuneva, M. O. et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 15, 157–170 (2012).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Kerr, E. M., Gaude, E., Turrell, F. K., Frezza, C. & Martins, C. P. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113 (2016).
Sciacovelli, M. & Frezza, C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 284, 3132–3144 (2017).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Ibrahimi, A. et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum. Gene Ther. 20, 845–860 (2009).
Agnese, S. T., Spierto, F. W. & Hannon, W. H. Evaluation of four reagents for delipidation of serum. Clin. Biochem. 16, 98–100 (1983).
Tumanov, S. et al. Calibration curve-free GC–MS method for quantitation of amino and non-amino organic acids in biological samples. Metabolomics 12, 1–13 (2016).
Mackay, G. M., Niels, L. Z., Broek, J. F. van den & Gottlieb, E. in Metabolic Analysis Using Stable Isotopes (ed., Metallo, C. M.) 171–196 (Elsevier, 2015).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Martin, M. & N, T. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Warnes, G. R. et al. gplots: Various R programming tools for plotting data. R package v.3.0-1. http://CRAN.R-project.org/package=gplots (2015).
Acknowledgements
We thank the patients and their families who generously donated the samples used in this study to the NHS Greater Glasgow and Clyde Biorepository, Laboratory Medicine Building, Queen Elizabeth University Hospital, the Bloodwise Childhood Leukemia Cell Bank, the Glasgow Neuroimmunology Biobank and the West of Scotland CSF Biobank. In addition, we thank J. Goodfellow, H. Willison, S. Bhatti and Y. Yousafzai for assistance with obtaining primary samples and C. Orange and L. Stevenson for help with histology. Histology slides were scanned by the University of Glasgow slide scanning and image analysis service at the Queen Elizabeth University Hospital. RNA-seq was performed by the Glasgow Polyomics research facility at the University of Glasgow. We also thank K. Keeshan and the Biological Services Unit, Cancer Research UK Beatson Institute for animal assistance. We thank G. Cazzaniga for supplying PDXs, V. Saha for providing reporter plasmids and L. Akimov, I. Muler, H. Fishman, A. Rein and E. Vax for technical assistance. This work was supported by the Chief Scientist Office (O.O. and C.H., grant ETM/374), Fondazione Italiana per la Ricerca sul Cancro *FIRC (A.M.S.), the William and Elizabeth Davies Foundation (A.C., Clinical Research Fellowship), the Laura and Ike Perlmutter Fund (E.G. and I.A.), the German Israel Foundation (S.I. and C.E.), the Norman and Sadie Lee Foundation (S.I.), the Israel Science Foundation 1775/12 (E.G. and I.M.), European Union ERA NET TRASCALL program (S.I.), Israel Cancer Research Foundation City of Hope collaborative program (S.I.) and Cancer Research UK (J.J.K. and G.M.). This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement META-CAN No 766214 (S.I.F., J.F-G., I.M. and E.G.).
Author information
Authors and Affiliations
Contributions
A.M.S., S.I.F., O.O., E.G., C.H., P.H. and S.I. designed the study. A.M.S., S.I.F., O.O., A.C., A.Z., S.B., I.G., L.F., Y.B., C.E., M.B., E.B. and S.J.R. provided the samples and performed most of the experiments. A.M.S., S.I.F., O.O., A.C., P.H., E.K.M., J.G.-F., C.H., I.M., J.R.C., M.G.K. and E.G. analyzed and interpreted the data. S.T., I.A., J.J.K. and G.M. performed and analyzed the MS experiments. A.M.S., S.I.F., O.O., C.H., E.G., I.M. and S.I. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
E.G. is a board member and a shareholder of Metabomed Ltd., Israel, J.J.K. is an employee and shareholder of Rheos Medicines Inc. M.G.K. is a consultant for Accent Therapeutics and M.G.K.’s laboratory receives some financial support from 28-7. These disclosures are not directly related to these studies. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Leukemic infiltrates in murine CNS and bone marrow.
a, H&E staining of paraffin-embedded leukemic (ALL) murine skull and brain. ** indicates leukemic infiltrate in calvarial bone marrow, dashed line indicates leptomeningeal space filled with leukemic infiltrate b, H&E staining of paraffin embedded murine femur confirming widespread dense leukemic infiltrate throughout the marrow cavity. Images are representative of 8 mice.
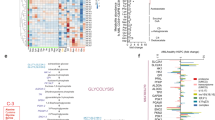
Extended Data Fig. 2 Fatty acid synthesis-related genes are upregulated in ALL cells derived from the CNS of xenograft models.
a, Schematic of the RNA Sequencing workflow. Two batches of five NSG mice were xenografted with human ALL cell lines SEM [t(4;11) MLL-AFF1 (MLL-AF4)]. Post engraftment, cells were collected from central nervous system (CNS) and spleens. Before RNA extraction, CNS or spleen ALL cells from each batch were pooled to reach the quantities required for polyA-tailed RNA sequencing. After extraction, RNA was sequenced and analyzed as described in material and methods. b, Top 20 differentially expressed coding genes in CNS compared to spleen from RNA sequencing, excluding genes with a base mean <10, ranked according to their adjusted p-value in SEM (I) and REH (II) cells. Gene function was assigned using NCBI Gene and linked resources. The reported p-value(s) result from a two-sided DESeq2’s Wald test and were FDR-adjusted by the Benjamini-Hochberg procedure. c, I-II Enrichment plots of metabolism of lipids and lipoproteins (REACTOME) and oxidative phosphorylation (KEGG) for REH cell line extracted from the CNS and spleen of engrafted mice (n=2 groups of 5 mice each). Profile of the running ES score & Positions of the Gene Set Members on the Rank Ordered List. III Statistically significant biological functions in REH cells isolated from CNS and spleen of xenografted mice. p-values for positive association with a signature (enrichment) were calculated by permutation test. Plotted are signatures with significant fold-changes in enrichment between the CNS versus spleen groups (log2 scale). Red bars indicate signatures with positive log fold-change (gain) in CNS versus spleen, blue bars indicate negative log fold-change (loss) in CNS versus spleen samples.
Extended Data Fig. 3 Schematic of the fatty acid metabolism.
Glucose or glutamine-derived citrate or free Acetyl-CoA serve as precursor for saturated fatty acid, further un-saturated to provide either triglycerides or phospholipids. Saturated fatty acids can also enter cycle of degradation within the mitochondrion through beta-oxidation. ACLY: ATP- Citrate Lyase; FASN: Fatty Acid Synthase; SCD: Stearoyl-CoA Desaturase; ACC: Acetyl CoA Carboxylase; CPT: Carnitine Palmitoyltransferase; HMGCR: 3-Hydroxy-3-Methylglutaryl-CoA Reductase; SQLE: Squalene epoxidase; TCA: Tricarboxylic acid cycle; FA: Fatty acid; FAO: Fatty acid oxidation.
Extended Data Fig. 4 Quantitative PCR validation of top ranked genes differentially expressed in CNS blasts compared to spleen in SEM and REH cells.
a, and 018z cells (b) p (two tailed) = one sample T and Wilcoxon test. Results are normalized to 36B4 human housekeeping genes and presented as LogFold2 change enrichment of comparing CNS to spleen for the SEM-REH samples; human HPRT was used as housekeeping gene and enrichment of comparing CNS to BM was calculated for 018z. n=7 for SCD, FASN, ACLY, CPT1a; n=6 for CPT1b; n=5 for CPT2 in (a). n=12 for LDLR, HMGCR, FASN, ACLY, CPT1a, CPT1b; n=11 for SQLE; n=7 for SCD, ABCA1 in b, For box-and-whisker plots, boxes represent 25th and 75th percentiles, center lines indicate median values and whiskers represent minimum and maximum values. c, Western blot of SCD and FASN proteins in SEM cells retrieved from the CNS and spleen of mice (n=4 mice).
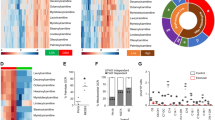
Extended Data Fig. 5 Analysis of available public human databases.
Left side of each panel: Boxplots showing single genes differentially regulated in samples of BM from patients at diagnosis (n=22) and relapse (n=20) and CNS samples at relapse (n=8) from public database GSE60926, unpaired analysis. Right side of each panel: Patient-derived xenograft samples established by transplantation of patient ALL cells onto NSG mice, single dots indicate paired bone marrow and CNS. Public database GSE89710. a, ABCA1: ATP-binding cassette transporter subfamily A member 1; b, ACC: Acetyl-CoA carboxylase; c, ACLY: ATP Citrate Lyase; d, CPT1A: Carnitine Palmitoyltransferase 1A; e, CPT1B: Carnitine Palmitoyltransferase 1B; f, CPT2: Carnitine Palmitoyltransferase 2; g, FASN: Fatty acid synthase; h, HMGCR: 3-Hydroxy-3-Methylglutaryl-CoA Reductase; i, LDLR: Low density lipoprotein receptor; j, SQLE: Squalene. FDR – false discovery rate. For box-and-whisker plots, boxes represent 25th and 75th percentiles, center lines indicate median values and whiskers represent minimum and maximum values.
Extended Data Fig. 6 Ratios of monounsaturated fatty acids to their saturated precursors.
Ratios of total levels of oleic/stearic acids in total fatty acids extracts in (a) SCD-high and (c) SCD-low 018z cells, in comparison to respective controls (CTL) (n=4 independent experiments for each condition). Ratios of relative levels of oleic/stearic acids in free fatty acids extracts in (b) 018z overexpressing or (d) downregulating SCD, comparatively to corresponding controls (CTL). n=5 independent experiments for each condition, p(two-tailed)=unpaired parametric Student’s t-test. Error bars represent mean ± s.d.
Extended Data Fig. 7 Increased SCD activity and expression upon genetic modification.
a, Ratio of relative level of oleic/stearic acids in free fatty acids extracts from cells isolated from CNS of mice engrafted with SCD overexpressing (SCD-high, n=3 mice) and control (CTL, n=4 mice) 018z cells. p(two-tailed)=unpaired parametric Student’s t-test. b, Gene expression level of SCD after overexpression in REH cells (n=2 independent experiments). Error bars represent mean ± s.d.
Extended Data Fig. 8 Effect of FBS delipidation on relative concentrations of metabolites, using fumed silica.
Relative levels of listed metabolites in lipidated (Lip) or delipidated (Delip) FBS – (a) stearic, (b) oleic, (c) palmitic, (d) palmitoleic, (e) arachidonic and (f) linoleic acids, (g) glucose, (h) lactate, (i) glutamine and (j) glutamate. Total fatty acids were extracted by saponification and the polar metabolites were extracted in 50% methanol, 30% acetonitrile, 20% water (“Reg extraction”).
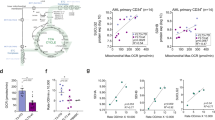
Extended Data Fig. 9 Body weight of mice treated with SCD1 inhibitor.
Body weight variation of NSG mice transplanted with PDX (a) 1, (b) 2, (c) 3, or (d) 4, treated with SW203668 (Treated, n=5 mice) or vehicle (Control, n=5 mice). Error bars represent mean ± s.d.
Extended Data Fig. 10 Example of the gating strategy used for flow cytometry analysis.
(a) Gating for live cells. (b) Gating to exclude doublets and cell aggregates. (c) Identification of human and mouse CD45+ specific populations.
Supplementary information
Supplementary Video
Phenotypic representation of SCD overexpression in ALL cells.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Savino, A.M., Fernandes, S.I., Olivares, O. et al. Metabolic adaptation of acute lymphoblastic leukemia to the central nervous system microenvironment depends on stearoyl-CoA desaturase. Nat Cancer 1, 998–1009 (2020). https://doi.org/10.1038/s43018-020-00115-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-020-00115-2
This article is cited by
-
Mass spectrometry-based proteomics of cerebrospinal fluid in pediatric central nervous system malignancies: a systematic review with meta-analysis of individual patient data
Fluids and Barriers of the CNS (2024)
-
Advances in regulation and function of stearoyl-CoA desaturase 1 in cancer, from bench to bed
Science China Life Sciences (2023)
-
Protamine 1 as a secreted colorectal cancer-specific antigen facilitating G1/S phase transition under nutrient stress conditions
Cellular Oncology (2023)
-
Transmembrane helices mediate the formation of a stable ternary complex of b5R, cyt b5, and SCD1
Communications Biology (2022)
-
Central nervous system involvement in childhood acute lymphoblastic leukemia is linked to upregulation of cholesterol biosynthetic pathways
Leukemia (2022)