Abstract
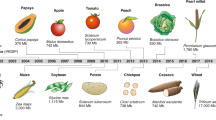
The global population continues to rise, as does the likelihood of reduced yields of major food crops due to the changing climate, thus making the development of genetically improved, stress-resilient crops a research priority. The convergence of low-cost genome sequencing with improved computational power and high-throughput molecular phenotyping technologies has accelerated the identification of genes underlying important agronomic traits relevant to food production and quality. Here, we discuss the evolution of plant improvement, and how researchers leverage genomic analyses and revolutionary new plant breeding technologies like site-directed nucleases to enhance food crop traits through agricultural biotechnology. Deployment of these products from the laboratory to the field remains hindered by biological and regulatory bottlenecks that require further development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Scott Camazine / Alamy Stock Photo (protein structure)

Similar content being viewed by others
References
Pingali, P. L. Green revolution: impacts, limits, and the path ahead. Proc. Natl Acad. Sci. USA 109, 12302–12308 (2012).
Wallace, J. G., Rodgers-Melnick, E. & Buckler, E. S. On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu. Rev. Genet. 52, 421–444 (2018).
Hunter, M. C., Smith, R. G., Schipanski, M. E., Atwood, L. W. & Mortensen, D. A. Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience 67, 386–391 (2017).
Tilman, D., Balzer, C., Hill, J. & Befort, B. L. Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA 108, 20260–20264 (2011).
Ray, D. K., Ramankutty, N., Mueller, N. D., West, P. C. & Foley, J. A. Recent patterns of crop yield growth and stagnation. Nat. Commun. 3, 1293 (2012).
Tietjen, B. et al. Climate change-induced vegetation shifts lead to more ecological droughts despite projected rainfall increases in many global temperate drylands. Glob. Chang. Biol. 23, 2743–2754 (2017).
Hirabayashi, Y. et al. Global flood risk underclimate change. Nat. Clim. Chang. 3, 816–821 (2013).
Rosenzweig, C. et al. Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc. Natl Acad. Sci. USA 111, 3268–3273 (2014).
Zhao, C. et al. Plausible rice yield losses under future climate warming. Nat. Plants 3, 16202 (2017).
Myers, S. S. et al. Increasing CO2 threatens human nutrition. Nature 510, 139–142 (2014).
Bebber, D. P., Ramotowski, M. A. T. & Gurr, S. J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 3, 985–988 (2013).
Meyer, R. S. & Purugganan, M. D. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013).
Olsen, K. M. & Wendel, J. F. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64, 47–70 (2013).
Hufford, M. B., Berny Mier y Teran, J. C. & Gepts, P. Crop biodiversity: an unfinished magnum opus of nature. Annu. Rev. Plant Biol. 70, 727–751 (2019).
Studer, A., Zhao, Q., Ross-Ibarra, J. & Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163 (2011).
Shang, Y. et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088 (2014).
Casañas, F., Simó, J., Casals, J. & Prohens, J. Toward an evolved concept of landrace. Front. Plant Sci. 8, 145 (2017).
Varshney, R. K. et al. Can genomics boost productivity of orphan crops? Nat. Biotechnol. 30, 1172–1176 (2012).
Gaut, B. S., Díez, C. M. & Morrell, P. L. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 31, 709–719 (2015).
Moyers, B. T., Morrell, P. L. & McKay, J. K. Genetic costs of domestication and improvement. J. Hered. 109, 103–116 (2018).
Oladosu, Y. et al. Principle and application of plant mutagenesis in crop improvement: a review. Biotechnol. Biotechnol. Equip. 30, 1–16 (2016).
Peng, J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999).
Rattanpal, H. S., Singh, G. & Gupta, M. Studies on mutation breeding in mandarin variety Kinnow. Curr. Sci. 116, 483–487 (2019).
Dwivedi, S. L. et al. Landrace germplasm for improving yield and abiotic stress adaptation. Trends Plant Sci. 21, 31–42 (2016).
Mickelbart, M. V., Hasegawa, P. M. & Bailey-Serres, J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251 (2015).
Xu, K. & Mackill, D. J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 2, 219–224 (1996).
Xu, K., Xu, X., Ronald, P. C. & Mackill, D. J. A high-resolution linkage map of the vicinity of the rice submergence tolerance locus Sub1. Mol. Gen. Genet. 263, 681–689 (2000).
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 (2006).
Ismail, A. M., Singh, U. S., Singh, S., Dar, M. H. & Mackill, D. J. The contribution of submergence-tolerant (Sub1) rice varieties to food security in flood-prone rainfed lowland areas in Asia. Field Crop. Res. 152, 83–93 (2013).
Emerick, K. & Ronald, P. C. Sub1 rice: engineering rice for climate change. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a034637 (2019).
Li, Z. et al. The 3,000 rice genomes project. Gigascience 3, 7 (2014).
Hendre, P. S. et al. African Orphan Crops Consortium (AOCC): status of developing genomic resources for African orphan crops. Planta 250, 989–1003 (2019).
Jupe, F. et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544 (2013).
Steuernagel, B. et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655 (2016).
Jouanin, A. et al. Development of the GlutEnSeq capture system for sequencing gluten gene families in hexaploid bread wheat with deletions or mutations induced by γ-irradiation or CRISPR/Cas9. J. Cereal Sci. 88, 157–166 (2019).
Tardieu, F., Cabrera-Bosquet, L., Pridmore, T. & Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 27, R770–R783 (2017).
Lee, T. et al. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 43, D996–D1002 (2015).
Brooks, M. D. et al. Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nat. Commun. 10, 1569 (2019).
Lee, I. et al. Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl Acad. Sci. USA 108, 18548–18553 (2011).
Friso, G. & Van Wijk, K. J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487 (2015).
McWhite, C. D. et al. A pan-plant protein complex map reveals deep conservation and novel assemblies. Cell 181, 460–474 (2020).
Huang, X. & Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 65, 531–551 (2014).
Zhu, G., Gou, J., Klee, H. & Huang, S. Next-gen approaches to flavor-related metabolism. Annu. Rev. Plant Biol. 70, 187–212 (2019).
Powell, A. L. T. et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715 (2012).
Zhu, G. et al. Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e12 (2018).
Bauchet, G. et al. Identification of major loci and genomic regions controlling acid and volatile content in tomato fruit: implications for flavor improvement. New Phytol. 215, 624–641 (2017).
Sauvage, C. et al. Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 165, 1120–1132 (2014).
Zhang, J. et al. Genome-wide association mapping for tomato volatiles positively contributing to tomato flavor. Front. Plant Sci. 6, 1042 (2015).
Tieman, D. et al. A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394 (2017).
Zhao, J. et al. Meta-analysis of genome-wide association studies provides insights into genetic control of tomato flavor. Nat. Commun. 10, 1534 (2019).
Crossa, J. et al. Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975 (2017).
Singh, A., Ganapathysubramanian, B., Singh, A. K. & Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124 (2016).
Zhang, Y., Malzahn, A. A., Sretenovic, S. & Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794 (2019).
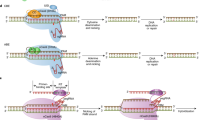
Puchta, H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14 (2005).
Huang, T.-K. & Puchta, H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 38, 443–453 (2019).
Hayut, S. F., Bessudo, C. M. & Levy, A. A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8, 15605 (2017).
Swinnen, G., Goossens, A. & Pauwels, L. Lessons from domestication: targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 21, 506–515 (2016).
Modrzejewski, D. et al. What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: a systematic map. Environ. Evid. 8, 27 (2019).
Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284 (2015).
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E. & Lippman, Z. B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8 (2017).
Weeks, D. P. Gene editing in polyploid crops: wheat, camelina, canola, potato, cotton, peanut, sugar cane, and citrus. Prog. Mol. Biol. Transl. Sci. 149, 65–80 (2017).
Dederer, H.-G. & Hamburger, D. (eds) Regulation of Genome Editing in Plant Biotechnology. Regulation of Genome Editing in Plant Biotechnology (Springer, 2019).
Breitler, J.-C. et al. CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. Plant Cell Tiss. Org. Cult. 134, 383–394 (2018).
Trevaskis, B. Developmental pathways are blueprints for designing successful crops. Front. Plant Sci. 9, 745 (2018).
Yang, Y. et al. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335 (2018).
Zheng, M. et al. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18, 644–654 (2019).
Zhou, J. et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 38, 475–485 (2019).
Huang, L. et al. Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. Crop J. 6, 475–481 (2018).
Li, M. et al. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7, 377 (2016).
Miao, C. et al. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl Acad. Sci. USA 115, 6058–6063 (2018).
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
Pavan, S., Jacobsen, E., Visser, R. G. F. & Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12 (2010).
van Schie, C. C. N. & Takken, F. L. W. Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581 (2014).
Nekrasov, V. et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482 (2017).
Wang, Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Fister, A. S., Landherr, L., Maximova, S. N. & Guiltinan, M. J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 9, 268 (2018).
Li, T., Liu, B., Spalding, M. H., Weeks, D. P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Oliva, R. et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350 (2019).
Peng, A. et al. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519 (2017).
Chandrasekaran, J. et al. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153 (2016).
Gomez, M. A. et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434 (2019).
Breiteneder, H. & Radauer, C. A classification of plant food allergens. J. Allergy Clin. Immunol. 113, 821–830 (2004).
Juhász, A. et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 4, eaar8602 (2018).
Sánchez-León, S. et al. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910 (2018).
García-Molina, M., Giménez, M., Sánchez-León, S. & Barro, F. Gluten free wheat: are we there? Nutrients 11, 487 (2019).
Dubois, A. E. J. et al. First successful reduction of clinical allergenicity of food by genetic modification: Mal d 1-silenced apples cause fewer allergy symptoms than the wild-type cultivar. Allergy Eur. J. Allergy Clin. Immunol. 70, 1406–1412 (2015).
Dodo, H. W., Konan, K. N., Chen, F. C., Egnin, M. & Viquez, O. M. Alleviating peanut allergy using genetic engineering: The silencing of the immunodominant allergen Ara h 2 leads to its significant reduction and a decrease in peanut allergenicity. Plant Biotechnol. J. 6, 135–145 (2008).
Wakasa, Y., Hirano, K., Urisu, A., Matsuda, T. & Takaiwa, F. Generation of transgenic rice lines with reduced contents of multiple potential allergens using a null mutant in combination with an RNA silencing method. Plant Cell Physiol. 52, 2190–2199 (2011).
Herman, E. M., Helm, R. M., Jung, R. & Kinney, A. J. Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 132, 36–43 (2003).
Li, X. et al. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9, 559 (2018).
Li, R. et al. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427 (2018).
Nonaka, S., Arai, C., Takayama, M., Matsukura, C. & Ezura, H. Efficient increase of γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7, 7057 (2017).
Zhang, H. et al. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898 (2018).
Li, A. et al. Editing of an alpha-kafirin gene family increases digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438 (2018).
Haun, W. et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940 (2014).
Morineau, C. et al. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15, 729–739 (2017).
Alexander, P. et al. Losses, inefficiencies and waste in the global food system. Agric. Syst. 153, 190–200 (2017).
Bachem, C. W. B. et al. Antisense expression of polyphenol oxidase genes inhibits enzymatic browning in potato tubers. Nat. Biotechnol. 12, 1101–1105 (1994).
Waltz, E. Nonbrowning GM apple cleared for market. Nature Biotechnology Blog http://blogs.nature.com/tradesecrets/2015/03/30/nonbrowning-gm-apple-cleared-for-market (2015).
Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293 (2016).
Re: Confirmation that PPO _ KO Potato is not a Regulated Article (United States Department of Agriculture, 2016); https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/16-090-01_air_response_signed.pdf
Re: Confirmation of the Regulatory Status of Genome edited Lactuca sativa (Lettuce) (United States Department of Agriculture, 2019); https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/18-243-01_air_response_signed.pdf
Non-browning lettuce is making its way to the market. AG Daily https://www.agdaily.com/news/non-browning-lettuce-market/ (3 June 2019).
Bruening, G. & Lyons, J. M. The case of the FLAVR SAVR tomato. Calif. Agric. 54, 6–7 (2000).
Wang, D. et al. Characterisation of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557 (2019).
Yang, L. et al. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 15, 1544–1555 (2017).
Uluisik, S. et al. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 34, 950–952 (2016).
Mieulet, D. et al. Turning rice meiosis into mitosis. Cell Res. 26, 1242–1254 (2016).
Wang, C. et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286 (2019).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019).
Østerberg, J. T. et al. Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci. 22, 373–384 (2017).
Li, T. et al. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163 (2018).
Zsögön, A. et al. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216 (2018).
Chopra, R. et al. Identification and stacking of crucial traits required for the domestication of pennycress. Nat. Food 1, 84–91 (2020).
Ribaut, J.-M. & Ragot, M. Modernising breeding for orphan crops: tools, methodologies, and beyond. Planta 250, 971–977 (2019).
Lemmon, Z. H. et al. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770 (2018).
Kwon, C., Heo, J., Lemmon, Z. H., Capua, Y. & Hutton, S. F. Rapid adaptation of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 38, 182–188 (2020).
Bull, S. E. et al. Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 4, eaat6086 (2018).
Jørgensen, K. et al. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol. 139, 363–374 (2005).
Tadele, Z. Orphan crops: their importance and the urgency of improvement. Planta 250, 677–694 (2019).
Fernie, A. R. & Yan, J. De novo domestication: an alternative route toward new crops for the future. Mol. Plant 12, 615–631 (2019).
Sunilkumar, G., Campbell, L. A. M., Puckhaber, L., Stipanovic, R. D. & Rathore, K. S. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl Acad. Sci. USA 103, 18054–18059 (2006).
Bradford, K. J., Van Deynze, A., Gutterson, N., Parrott, W. & Strauss, S. H. Regulating transgenic crops sensibly: lessons from plant breeding, biotechnology and genomics. Nat. Biotechnol. 23, 439–444 (2005).
Napier, J. A., Haslam, R. P., Tsalavouta, M. & Sayanova, O. The challenges of delivering genetically modified crops with nutritional enhancement traits. Nat. Plants 5, 563–567 (2019).
Brief 54: Global Status of Commercialized Biotech/GM Crops in 2018 (ISAAA, 2018).
GM Approval Database (ISAAA, accessed 1 October 2019); http://www.isaaa.org/gmapprovaldatabase/
Klümper, W. & Qaim, M. A meta-analysis of the impacts of genetically modified crops. PLoS One 9, e111629 (2014).
Dively, G. P. et al. Regional pest suppression associated with widespread Bt maize adoption benefits vegetable growers. Proc. Natl Acad. Sci. USA 115, 3320–3325 (2018).
Kouser, S. & Qaim, M. Impact of Bt cotton on pesticide poisoning in smallholder agriculture: a panel data analysis. Ecol. Econ. 70, 2105–2113 (2011).
Wu, F. Mycotoxin reduction in Bt corn: Potential economic, health, and regulatory impacts. Transgenic Res. 15, 277–289 (2006).
Shelton, A. M. et al. Bt brinjal in Bangladesh: the first genetically engineered food crop in a developing country. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a034678 (2019).
Kawashima, C. G. et al. A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 34, 661–665 (2016).
La Concepcion, J. D., Franceschetti, M., Terauchi, R., Kamoun, S. & Banfield, M. Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife 8, e47713 (2019).
Wurtzel, E. T. et al. Revolutionizing agriculture with synthetic biology. Nat. Plants 5, 1207–1210 (2019).
South, P. F., Cavanagh, A. P., Liu, H. W. & Ort, D. R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077 (2019).
Dalal, J. et al. A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa. Biotechnol. Biofuels 8, 175 (2015).
Manna, M., Achary, V. M. M., Islam, T., Agrawal, P. K. & Reddy, M. K. The development of a phosphite-mediated fertilization and weed control system for rice. Sci. Rep. 6, 24942 (2016).
Pandeya, D. et al. Selective fertilization with phosphite allows unhindered growth of cotton plants expressing the ptxD gene while suppressing weeds. Proc. Natl Acad. Sci. USA 115, E6946–E6955 (2018).
Mus, F. et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 82, 3698–3710 (2016).
Ye, X. & Beyer, P. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305 (2000).
Paine, J. A. et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487 (2005).
Paul, J.-Y. et al. Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 15, 520–532 (2017).
Zhu, Q. et al. From Golden Rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol. Plant 11, 1440–1448 (2018).
Dong, O. X. et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 11, 1178 (2020).
Ainley, W. M. et al. Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134 (2013).
Sun, Y., Li, J. & Xia, L. Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front. Plant Sci. 7, 1928 (2016).
Qi, Y. et al. Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res. 23, 547–554 (2013).
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A. & Voytas, D. F. DNA replicons for plant genome engineering. Plant Cell 26, 151–63 (2014).
Dahan-Meir, T. et al. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95, 5–16 (2018).
Gil-Humanes, J. et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89, 1251–1262 (2017).
Altpeter, F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016).
Camacho, A., Van Deynze, A., Chi-Ham, C. & Bennett, A. B. Genetically engineered crops that fly under the US regulatory radar. Nat. Biotechnol. 32, 1087–1091 (2014).
McHughen, A. & Smyth, S. US regulatory system for genetically modified [genetically modified organism (GMO), rDNA or transgenic] crop cultivars. Plant Biotechnol. J. 6, 2–12 (2008).
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Mark Cigan, A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274 (2016).
Liu, J. et al. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31, 368–383 (2019).
Jupe, F. et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 15, e1007819 (2019).
Lowe, K. et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015 (2016).
Lowe, K. et al. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54, 240–252 (2018).
Maher, M. F. et al. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89 (2019).
Larkin, P. J. & Scowcroft, W. R. Somaclonal variation — a novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 60, 197–214 (1981).
Tang, X. et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19, 84 (2018).
Li, J. et al. Whole genome sequencing reveals rare off‐target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR /Cas9‐edited cotton plants. Plant Biotechnol. J. 17, 858–868 (2019).
Fossi, M., Amundson, K., Kuppu, S., Britt, A. & Comai, L. Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86 (2019).
Han, Z. et al. Heritable epigenomic changes to the maize methylome resulting from tissue culture. Genetics 209, 983–995 (2018).
Lee, K. & Seo, P. J. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 23, 235–247 (2018).
Stroud, H. et al. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2, e00354 (2013).
Wibowo, A. et al. Partial maintenance of organ-specific epigenetic marks during plant asexual reproduction leads to heritable phenotypic variation. Proc. Natl Acad. Sci. USA 115, E9145–E9152 (2018).
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L. & Landry, M. P. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018).
Demirer, G. S. et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 14, 456–464 (2019).
Kwak, S.-Y. et al. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 14, 447–455 (2019).
Zhao, X. et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3, 956–964 (2017).
Kelliher, T. et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 37, 287–292 (2019).
Wang, B. et al. Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 12, 597–602 (2019).
Miller, J. K. & Bradford, K. J. The regulatory bottleneck for biotech specialty crops. Nat. Biotechnol. 28, 1012–1014 (2010).
National Organic Standards Board Materials/GMO Subcommittee Proposal Excluded Methods Determinations (2019); https://www.ams.usda.gov/sites/default/files/media/MSExcludedMethodsProposaFall2019.pdf
National Organic Program Policy Memorandum 13 (USDA, 2013); https://www.ams.usda.gov/sites/default/files/media/NOP-PM-13-1-CellFusion.pdf
Ishii, T. & Araki, M. Consumer acceptance of food crops developed by genome editing. Plant Cell Rep. 35, 1507–1518 (2016).
Urnov, F. D., Ronald, P. C. & Carroll, D. A call for science-based review of the European court’s decision on gene-edited crops. Nat. Biotechnol. 36, 800–802 (2018).
Faure, J. D. & Napier, J. A. Europe’s first and last field trial of gene-edited plants? eLife 7, e42379 (2018).
Secretary Perdue Issues USDA Statement on Plant Breeding Innovation USDA Press Release https://www.usda.gov/media/press-releases/2018/03/28/secretary-perdue-issues-usda-statement-plant-breeding-innovation (2018).
Waltz, E. With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36, 6–7 (2018).
Gheysen, G., Custers, R., Gheysen, G. & Custers, R. Why organic farming should embrace co-existence with cisgenic late blight–resistant potato. Sustainability 9, 172 (2017).
Husaini, A. M. & Sohail, M. Time to redefine organic agriculture: can’t GM crops be certified as organics? Front. Plant Sci. 9, 423 (2018).
Andersen, M. M. et al. Feasibility of new breeding techniques for organic farming. Trends Plant Sci. 20, 426–434 (2015).
Scheufele, D. A. & Krause, N. M. Science audiences, misinformation, and fake news. Proc. Natl Acad. Sci. USA 116, 7662–7669 (2019).
Hilscher, J., Bürstmayr, H. & Stoger, E. Targeted modification of plant genomes for precision crop breeding. Biotechnol. J. 12, 1600173 (2017).
Acknowledgements
We apologize to those authors whose research could not be cited due to space limits. We thank H. Bartram for a careful reading of the manuscript. M.A.S. was supported by the Corteva Agriscience Open Innovation programme grant entitled “Gene Editing for Organic Agriculture.” P.C.R. was supported by grants from the US National Science Foundation (award no. 1237975), the Crary Social Ecology Fund, the Foundation for Food and Agricultural Research (award no. 534683) and the National Institutes of Health (GM122968).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Steinwand, M.A., Ronald, P.C. Crop biotechnology and the future of food. Nat Food 1, 273–283 (2020). https://doi.org/10.1038/s43016-020-0072-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43016-020-0072-3
This article is cited by
-
Can plant hormonomics be built on simple analysis? A review
Plant Methods (2023)
-
Osa-miR1861e Modulates Sodium Toxicity Responses in Rice Immature Grains via OsGST and OsPILS7b
Journal of Plant Growth Regulation (2023)
-
Understanding the Origin and Evolution of Tea (Camellia sinensis [L.]): Genomic Advances in Tea
Journal of Molecular Evolution (2023)
-
Target-oriented prioritization: targeted selection strategy by integrating organismal and molecular traits through predictive analytics in breeding
Genome Biology (2022)
-
Estimating maize seedling number with UAV RGB images and advanced image processing methods
Precision Agriculture (2022)