Abstract
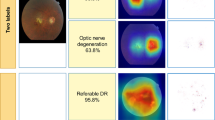
Ultra-widefield (UWF) imaging is a promising modality that captures a larger retinal field of view compared with traditional fundus photography. Previous studies have shown that deep learning models are effective for detecting retinal disease in UWF images, but primarily considered individual diseases under less-than-realistic conditions (excluding images with other diseases, artefacts, comorbidities or borderline cases; and balancing healthy and diseased images) and did not systematically investigate which regions of the UWF images are relevant for disease detection. Here we first improve on the state of the field by proposing a deep learning model that can recognize multiple retinal diseases under more realistic conditions than what has previously been considered. We then use global explainability methods to identify which regions of the UWF images the model generally attends to. Our model performs very well, separating between healthy and diseased retinas with an area under the receiver operating characteristic curve (AUC) of 0.9196 (±0.0001) on an internal test set, and an AUC of 0.9848 (±0.0004) on a challenging, external test set. When diagnosing specific diseases, the model attends to regions where we would expect those diseases to occur. We further identify the posterior pole as the most important region in a purely data-driven fashion. Surprisingly, 10% of the image around the posterior pole is sufficient for achieving comparable performance across all labels to having the full images available.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data are available from Hitoshi Tabuchi and the other authors of the Tsukazaki Optos Public Project subject to current export restrictions, which are imposed by Japanese legislation at the time of writing. Previously, it was publicly accessible via a project website where we obtained the copy used in this study. A subset containing images images of healthy eyes and eyes with RP used in a previous study15 is publicly accessible directly online at https://figshare.com/authors/Masahiro_Kameoka/6020591. The external validation set we assembled from the American Society of Retina Specialists Retina Image Bank (https://imagebank.asrs.org), RetinaRocks Image Library (https://www.retinarocks.org/) and Optos Recognising Pathology resource (https://recognizingpathology.optos.com/) is described in Supplementary Section 3 in sufficient detail to reproduce the dataset. We also note that the dataset is well known within the community (for example, refs. 16,26).
Code availability
The code for this project, a requirements.txt file listing all libraries used and their versions, and the trained model are available online at https://github.com/justinengelmann/UWF_multiple_disease_detection.
References
Brown, G. C. Vision and quality-of-life. Trans. Am. Ophthalmol.Soc. 97, 473–511 (1999).
Pezzullo, L., Streatfeild, J., Simkiss, P. & Shickle, D. The economic impact of sight loss and blindness in the UK adult population. BMC Health Serv. Res. 18, 63 (2018).
Patel, S. N., Shi, A., Wibbelsman, T. D. & Klufas, M. A. Ultra-widefield retinal imaging: an update on recent advances. Ther. Adv. Ophthalmol. 12, https://journals.sagepub.com/doi/10.1177/2515841419899495 (2020).
Nagiel, A., Lalane, R. A., Sadda, S. R. & Schwartz, S. D. Ultra-widefield fundus imaging: a review of clinical applications and future trends. Retina 36, 660–678 (2016).
Tan, T.-E., Ting, D. S. W., Wong, T. Y. & Sim, D. A. Deep learning for identification of peripheral retinal degeneration using ultra-wide-field fundus images: is it sufficient for clinical translation? Ann. Transl. Med. 8, 611 (2020).
Matsuba, S. et al. Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int. Ophthalmol. 39, 1269–1275 (2019).
Nagasato, D. et al. Deep-learning classifier with ultrawide-field fundus ophthalmoscopy for detecting branch retinal vein occlusion. Int. J. Ophthalmol. 12, 94–99 (2019).
Nagasato, D. et al. Deep neural network-based method for detecting central retinal vein occlusion using ultrawide-field fundus ophthalmoscopy. J. Ophthalmol. 2018, 1875431 (2018).
Tabuchi, H., Masumoto, H., Nakakura, S., Noguchi, A. & Tanabe, H. Discrimination ability of glaucoma via DCNNs models from ultra-wide angle fundus images comparing either full or confined to the optic disc. In Asian Conference on Computer Vision 229–234 (Springer, 2018).
Masumoto, H. et al. Deep-learning classifier with an ultrawide-field scanning laser ophthalmoscope detects glaucoma visual field severity. J. Glaucoma 27, 647–652 (2018).
Nagasawa, T. et al. Accuracy of deep learning, a machine learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting idiopathic macular holes. PeerJ 6, e5696 (2018).
Nagasawa, T. et al. Accuracy of ultrawide-field fundus ophthalmoscopy-assisted deep learning for detecting treatment-naive proliferative diabetic retinopathy. Int. Ophthalmol. 39, 2153–2159 (2019).
Ohsugi, H., Tabuchi, H., Enno, H. & Ishitobi, N. Accuracy of deep learning, a machine-learning technology, using ultra–wide-field fundus ophthalmoscopy for detecting rhegmatogenous retinal detachment. Sci. Rep. 7, 9425 (2017).
Masumoto, H. et al. Retinal detachment screening with ensembles of neural network models. In Asian Conference on Computer Vision 251–260 (Springer, 2018).
Masumoto, H. et al. Accuracy of a deep convolutional neural network in detection of retinitis pigmentosa on ultrawide-field images. PeerJ 7, e6900 (2019).
Antaki, F. et al. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br. J. Ophthalmol. https://bjo.bmj.com/content/early/2021/08/02/bjophthalmol-2021-319030.info (2021).
Hemelings, R. et al. Deep learning on fundus images detects glaucoma beyond the optic disc. Sci. Rep. 11, 20313 (2021).
Duker, J. S. et al. The international vitreomacular traction study group classification of vitreomacular adhesion, traction, and macular hole. Ophthalmology 120, 2611–2619 (2013).
Beede, E. et al. A human-centered evaluation of a deep learning system deployed in clinics for the detection of diabetic retinopathy. In Proc. 2020 CHI Conference on Human Factors in Computing Systems 1–12 (Association for Computing Machinery, 2020).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. In Proc. IEEE International Conference on Computer Vision 618–626 (IEEE, 2017).
Engelmann, J., Storkey, A. & Bernabeu, M. O. Global explainability in aligned image modalities. Preprint at https://arxiv.org/abs/2112.09591 (2021).
Wilkinson, C. P., Hinton, D. R., Sadda, S. R. & Wiedemann, P. Ryan’s Retina 6th edn (Elsevier Health Sciences, 2018).
DeGrave, A. J., Janizek, J. D. & Lee, S.-I. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 3, 610–619 (2021).
Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Yamashita, T. et al. Factors in color fundus photographs that can be used by humans to determine sex of individuals. Transl. Vis. Sci. Technol. 9, 4 (2020).
Khan, S. M. et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit. Health 3, e51–e66 (2021).
González-Gonzalo, C., Liefers, B., van Ginneken, B. & Sánchez, C. I. Iterative augmentation of visual evidence for weakly-supervised lesion localization in deep interpretability frameworks: application to color fundus images. IEEE Tran. Med. Imaging 39, 3499–3511 (2020).
Quellec, G., Charrière, K., Boudi, Y., Cochener, B. & Lamard, M. Deep image mining for diabetic retinopathy screening. Med. Image Anal. 39, 178–193 (2017).
Zhang, H., Cisse, M., Dauphin, Y. N. & Lopez-Paz, D. mixup: beyond empirical risk minimization. Preprint at https://arxiv.org/abs/1710.09412 (2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning 448–456 (PMLR, 2015).
Wightman, R., Touvron, H. & Jégou, H. ResNet strikes back: an improved training procedure in timm. Preprint at https://arxiv.org/abs/2110.00476 (2021).
Bello, I. et al. Revisiting ResNets: improved training and scaling strategies. Preprint at https://arxiv.org/abs/2103.07579 (2021).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In Proc. IEEE International Conference on Computer Vision 1026–1034 (IEEE, 2015).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2009).
Loshchilov, I. & Hutter, F. SGDR: stochastic gradient descent with warm restarts. Preprint at https://arxiv.org/abs/1608.03983 (2016).
Izmailov, P., Podoprikhin, D., Garipov, T., Vetrov, D. & Wilson, A. G. Averaging weights leads to wider optima and better generalization. Preprint at https://arxiv.org/abs/1803.05407 (2018).
Zhong, Z., Zheng, L., Kang, G., Li, S. & Yang, Y. Random erasing data augmentation. In Proc. AAAI Conference on Artificial Intelligence Vol. 34, 13001–13008 (Association for the Advancement of Artificial Intelligence, 2020).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2818–2826 (IEEE, 2016).
Müller, R., Kornblith, S. & Hinton, G. When does label smoothing help? Preprint at https://arxiv.org/abs/1906.02629 (2019).
Krause, J. et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology 125, 1264–1272 (2018).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Friedman, J. et al. The Elements of Statistical Learning Vol. 1 (Springer Series in Statistics, Springer, 2001).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8024–8035 (2019).
Wightman, R. PyTorch image models. GitHub https://github.com/rwightman/pytorch-image-models (2019).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sc. Eng. 9, 90–95 (2007).
Waskom, M. L. seaborn: statistical data visualization. J. Open Source Soft. 6, 3021 (2021).
Acknowledgements
We thank H. Masumoto and H. Tabuchi as well as D. Nagasato, S. Nakakura, M. Kameoka, R. Aoki, T. Sogawa, S. Matsuba, H. Tanabe, T. Nagasawa, Y. Yoshizumi, T. Sonobe, T. Yamauchi and all their colleagues at Tsukazaki Hospital for releasing the TOP dataset. This is a great contribution to AI research in ophthalmology for which we are most grateful. We also thank the American Society of Retina Specialists for their Retina Image Bank, and RetinaRocks for their Image Library. We further thank all users that submitted images for research use to these online repositories or elsewhere. This work was supported by the United Kingdom Research and Innovation (grant EP/S02431X/1), UKRI Centre for Doctoral Training in Biomedical AI at the University of Edinburgh, School of Informatics. For the purpose of open access, the author has applied a creative commons attribution (CC BY) licence to any author accepted manuscript version arising. This work was supported by The Royal College of Surgeons of Edinburgh, Sight Scotland, The RS Macdonald Charitable Trust, Chief Scientist Office, and Edinburgh & Lothians Health Foundation through a proof-of-concept award for the SCONe project. Grant EP/S02431X/1: J.E. SCONe project grants: A.D.M. and E.P.
Author information
Authors and Affiliations
Contributions
J.E. was responsible for all aspects of this work, including conceptualization, study design/methods, experiments, analysis, interpretation, figures and writing. A.S. and M.O.B. jointly supervised and contributed to all aspects of this work. A.D.M., I.J.C.M. and E.P. provided domain expertise regarding ophthalmology and ultra-widefield imaging, assessed the top 20 false positives, and provided feedback on the interpretation of the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Edward Korot and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Six of nine images showing the same eye of the same patient.
Six of nine images showing the same eye of the same patient. All images show DR according to the labels. While there are some differences between the images in terms of artefacts and pathology, the general pattern of the pathology is consistent between images and could be memorized by a model.
Supplementary information
Supplementary Information
Supplementary Information, containing all supplementary sections (text, figures and tables).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Engelmann, J., McTrusty, A.D., MacCormick, I.J.C. et al. Detecting multiple retinal diseases in ultra-widefield fundus imaging and data-driven identification of informative regions with deep learning. Nat Mach Intell 4, 1143–1154 (2022). https://doi.org/10.1038/s42256-022-00566-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00566-5
This article is cited by
-
Enhancing rare retinal disease classification: a few-shot meta-learning framework utilizing fundus images
Multimedia Tools and Applications (2023)
-
Retinal disease prediction through blood vessel segmentation and classification using ensemble-based deep learning approaches
Neural Computing and Applications (2023)