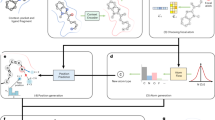
Abstract
Over the past decade, the illicit drug market has been reshaped by the proliferation of clandestinely produced designer drugs. These agents, referred to as new psychoactive substances (NPSs), are designed to mimic the physiological actions of better-known drugs of abuse while skirting drug control laws. The public health burden of NPS abuse obliges toxicological, police and customs laboratories to screen for them in law enforcement seizures and biological samples. However, the identification of emerging NPSs is challenging due to the chemical diversity of these substances and the fleeting nature of their appearance on the illicit market. Here we present DarkNPS, a deep learning-enabled approach to automatically elucidate the structures of unidentified designer drugs using only mass spectrometric data. Our method employs a deep generative model to learn a statistical probability distribution over unobserved structures, which we term the structural prior. We show that the structural prior allows DarkNPS to elucidate the exact chemical structure of an unidentified NPS with an accuracy of 51% and a top-10 accuracy of 86%. Our generative approach has the potential to enable de novo structure elucidation for other types of small molecules that are routinely analysed by mass spectrometry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
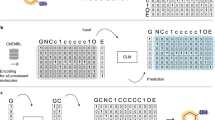
Owing to the sensitivity of the data and the potential for misuse, HighResNPS and the databases of generated molecules and MS/MS spectra described here are not available to the public for unrestricted download. However, the data have been uploaded to the NPS Data Hub59 (https://nps-datahub.com/) and will be made available to all qualified researchers in the field upon request. A demonstration dataset of 2,000 SMILES strings for drug-like small molecules sampled at random from the ChEMBL database is provided at http://github.com/skinnider/NPS-generation to demonstrate the functionality of the code.
Code availability
Code used to train and evaluate chemical language models is available from GitHub at http://github.com/skinnider/NPS-generation (https://doi.org/10.5281/zenodo.5136669).
References
Peacock, A. et al. New psychoactive substances: challenges for drug surveillance, control and public health responses. Lancet 394, 1668–1684 (2019).
Baumann, M. H. et al. Baths salts, spice and related designer drugs: the science behind the headlines. J. Neurosci. 34, 15150–15158 (2014).
Underwood, E. A new drug war. Science 347, 469–473 (2015).
Brandt, S. D., King, L. A. & Evans-Brown, M. The new drug phenomenon. Drug Test. Anal. 6, 587–597 (2014).
Nichols, D. Legal highs: the dark side of medicinal chemistry. Nature 469, 7 (2011).
Bijlsma, L. et al. Mass spectrometric identification and structural analysis of the third-generation synthetic cannabinoids on the UK market since the 2013 legislative ban. Forensic Toxiocol. 35, 376–388 (2017).
Baumann, M. H. & Volkow, N. D. Abuse of new psychoactive substances: threats and solutions. Neuropsychopharmacology 41, 663–665 (2016).
Johnson, L. A., Johnson, R. L. & Portier, R.-B. Current ‘legal highs’. J. Emerg. Med. 44, 1108–1115 (2013).
Luciano, R. L. & Perazella, M. A. Nephrotoxic effects of designer drugs: synthetic is not better! Nat. Rev. Nephrol. 10, 314–324 (2014).
Gebel Berg, E. Designer drug detective work. ACS Cent. Sci. 2, 363–366 (2016).
Carroll, F. I., Lewin, A. H., Mascarella, S. W., Seltzman, H. H. & Reddy, P. A. Designer drugs: a medicinal chemistry perspective. Ann. N. Y. Acad. Sci. 1248, 18–38 (2012).
Lewin, A. H., Seltzman, H. H., Carroll, F. I., Mascarella, S. W. & Reddy, P. A. Emergence and properties of spice and bath salts: a medicinal chemistry perspective. Life Sci. 97, 9–19 (2014).
von Cüpper, M., Dalsgaard, P. W. & Linnet, K. Identification of new psychoactive substances in seized material using UHPLC-QTOF-MS and an online mass spectral database. J. Anal. Toxicol. 44, 1047–1051 (2021).
Firman, J. W. et al. Chemoinformatic consideration of novel psychoactive substances: compilation and preliminary analysis of a categorised dataset. Mol. Inform. 38, e1800142 (2019).
Mardal, M. et al. HighResNPS.com: an online crowd-sourced HR-MS database for suspect and non-targeted screening of new psychoactive substances. J. Anal. Toxicol. 43, 520–527 (2019).
Wohlfarth, A. & Weinmann, W. Bioanalysis of new designer drugs. Bioanalysis 2, 965–979 (2010).
Bell, C., George, C., Kicman, A. T. & Traynor, A. Development of a rapid LC-MS/MS method for direct urinalysis of designer drugs. Drug Test. Anal. 3, 496–504 (2011).
Pasin, D., Cawley, A., Bidny, S. & Fu, S. Current applications of high-resolution mass spectrometry for the analysis of new psychoactive substances: a critical review. Anal. Bioanal. Chem. 409, 5821–5836 (2017).
Reitzel, L. A., Dalsgaard, P. W., Müller, I. B. & Cornett, C. Identification of ten new designer drugs by GC-MS, UPLC-QTOF-MS and NMR as part of a police investigation of a Danish internet company. Drug Test. Anal. 4, 342–354 (2012).
Tsochatzis, E. et al. Identification of 1-butyl-lysergic acid diethylamide (1B-LSD) in seized blotter paper using an integrated workflow of analytical techniques and chemo-informatics. Molecules 25, 712 (2020).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Gómez-Bombarelli, R. et al. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat. Mater. 15, 1120–1127 (2016).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018).
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminform. 9, 48 (2017).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: generative models for matter engineering. Science 361, 360–365 (2018).
Elton, D. C., Boukouvalas, Z., Fuge, M. D. & Chung, P. W. Deep learning for molecular design—a review of the state of the art. Mol. Syst. Des. Eng. 4, 828–849 (2019).
Skinnider, M. A., Stacey, R. G., Wishart, D. S. & Foster, L. J. Chemical language models enable navigation in sparsely populated chemical space. Nat. Mach. Intell. 3, 759–770 (2021).
Bjerrum, E. J. SMILES enumeration as data augmentation for neural network modeling of molecules. Preprint at https://arxiv.org/abs/1703.07076 (2017).
Scheubert, K., Hufsky, F. & Böcker, S. Computational mass spectrometry for small molecules. J. Cheminform. 5, 12 (2013).
Bertz, S. H. The first general index of molecular complexity. J. Am. Chem. Soc. 103, 3599–3601 (1981).
Wildman, S. A. & Crippen, G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 39, 868–873 (1999).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Ertl, P., Roggo, S. & Schuffenhauer, A. Natural product-likeness score and its application for prioritization of compound libraries. J. Chem. Inf. Model. 48, 68–74 (2008).
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 1, 8 (2009).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Shaker, B. et al. LightBBB: computational prediction model of blood–brain-barrier penetration based on LightGBM. Bioinformatics 37, 1135–1139 (2021).
Bajusz, D., Rácz, A. & Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 7, 20 (2015).
Skinnider, M. A., Dejong, C. A., Franczak, B. C., McNicholas, P. D. & Magarvey, N. A. Comparative analysis of chemical similarity methods for modular natural products with a hypothetical structure enumeration algorithm. J. Cheminform. 9, 46 (2017).
Preuer, K., Renz, P., Unterthiner, T., Hochreiter, S. & Klambauer, G. Fréchet ChemNet distance: a metric for generative models for molecules in drug discovery. J. Chem. Inf. Model. 58, 1736–1741 (2018).
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Blaženović, I. et al. Comprehensive comparison of in silico MS/MS fragmentation tools of the CASMI contest: database boosting is needed to achieve 93% accuracy. J. Cheminform. 9, 32 (2017).
Skinnider, M. A. et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11, 6058 (2020).
Winter, R., Montanari, F., Noé, F. & Clevert, D.-A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 10, 1692–1701 (2019).
Allen, F., Greiner, R. & Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 11, 98–110 (2015).
Djoumbou-Feunang, Y. et al. CFM-ID 3.0: significantly improved ESI-MS/MS prediction and compound identification. Metabolites 9, 72 (2019).
Renz, P., Van Rompaey, D., Wegner, J. K., Hochreiter, S. & Klambauer, G. On failure modes in molecule generation and optimization. Drug Discov. Today Technol. 32–33, 55–63 (2019).
Brown, N., Fiscato, M., Segler, M. H. S. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019).
Jonas, E. & Kuhn, S. Rapid prediction of NMR spectral properties with quantified uncertainty. J. Cheminform. 11, 50 (2019).
Kwon, Y., Lee, D., Choi, Y.-S., Kang, M. & Kang, S. Neural message passing for NMR chemical shift prediction. J. Chem. Inf. Model. 60, 2024–2030 (2020).
Cobas, C. NMR signal processing, prediction and structure verification with machine learning techniques. Magn. Reson. Chem. 58, 512–519 (2020).
Moret, M., Friedrich, L., Grisoni, F., Merk, D. & Schneider, G. Generative molecular design in low data regimes. Nat. Mach. Intell. 2, 171–180 (2020).
Arús-Pous, J. et al. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminform. 11, 71 (2019).
Blaschke, T. et al. REINVENT 2.0: an AI tool for de novo drug design. J. Chem. Inf. Model. 60, 5918–5922 (2020).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
O’Boyle, N. M. & Sayle, R. A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminform. 8, 36 (2016).
Riniker, S. & Landrum, G. A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 5, 26 (2013).
Böcker, S. Searching molecular structure databases using tandem MS data: are we there yet? Curr. Opin. Chem. Biol. 36, 1–6 (2017).
Urbas, A. et al. NPS data hub: a web-based community driven analytical data repository for new psychoactive substances. Forensic Chem. 9, 76–81 (2018).
Acknowledgements
This work was supported by funding from Genome Canada, Genome British Columbia and Genome Alberta (project 284MBO), the National Institutes of Health (NIH), National Institute of Environmental Health Sciences grant no. U2CES030170 and computational resources provided by WestGrid, Compute Canada and Advanced Research Computing at the University of British Columbia. M.A.S. acknowledges support from a CIHR Vanier Canada Graduate Scholarship, a Roman M. Babicki Fellowship in Medical Research, a Borealis AI Graduate Fellowship, a Walter C. Sumner Memorial Fellowship and a Vancouver Coastal Health–CIHR–UBC MD/PhD Studentship.
Author information
Authors and Affiliations
Contributions
All authors contributed to study design. M.A.S. and F.W. performed experiments. P.W.D. supervised the analysis of DXME at RKA. All authors contributed to data analysis. M.A.S. wrote the manuscript. All authors edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Claude Guillou, Stefan Kuhn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Model selection and hyperparameter optimization.
a, Jensen-Shannon distance between the distribution of Murcko scaffolds in the training set and generated molecules, for recurrent neural network-based models trained on the HighResNPS database after varying degrees of non-canonical SMILES enumeration. b, Jensen-Shannon distance between the natural product-likeness scores of the training set and generated molecules, for recurrent neural network-based models trained on the HighResNPS database after varying degrees of non-canonical SMILES enumeration. c, Jensen-Shannon distance between the proportion of stereocenters in the training set and generated molecules, for recurrent neural network-based models trained on the HighResNPS database after varying degrees of non-canonical SMILES enumeration. d, Factor loadings onto the first principal component in a principal component analysis of recurrent neural network-based models trained on the HighResNPS database after varying degrees of non-canonical SMILES enumeration.
Extended Data Fig. 2 Physicochemical properties and EMCDDA drug categorizations of generated molecules.
a, Calculated octanol-water partition coefficients (LogP) of known NPSs and generated molecules. b, Topological complexities of known NPSs and generated molecules. c, Natural product-likeness scores of known NPSs and generated molecules. d, Synthetic accessibility scores of known NPSs and generated molecules. e, UMAP visualization of known NPSs and an equal number of generated molecules sampled at random from the trained generative model, with the known NPSs colored by their EMCDDA drug categorizations. f, Log-odds ratios of EMCDDA drug category frequencies among generated molecules, as compared to the training set. *, p < 0.05; ***, p < 0.001.
Extended Data Fig. 3 Sampling frequencies of known and generated molecules.
a, Distribution of sampling frequencies within a sample of 1 billion SMILES strings from the trained generative model, with known NPSs from the training set shown in red. b, Jensen-Shannon distance between the molecular weights of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. c, Jensen-Shannon distance between the quantitative estimate of drug-likeness (QED) score of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. d, Jensen-Shannon distance between the proportion of carbons that are sp3-hybridized in generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. e, Jensen-Shannon distance between the partition coefficients of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. f, Jensen-Shannon distance between the topological complexities of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. g, Jensen-Shannon distance between the natural product-likeness scores of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. h, Jensen-Shannon distance between the synthetic accessibility scores of generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies. i, Jensen-Shannon distance between the proportion of stereocenters in generated molecules and the set of known NPSs, for molecules generated with progressively increasing frequencies.
Extended Data Fig. 4 Examples of molecules from the held-out set that were not generated by DarkNPS.
Chemical structures of an illustrative subset of the 18 molecules in the held-out set that were never produced by the generative model in a sample of 1 billion SMILES strings, and their nearest neighbors among structures that were generated by the model. Many of these molecules either are not designer drugs at all (for example, clozapine, citicoline, nalmefene, 2,2-dibromo-1-phenylhexan-2-one), or had a very closely related molecule appear in the model output.
Extended Data Fig. 5 Examples of molecules from the held-out set that were correctly anticipated by DarkNPS.
a, Frequency with which each of the 194 molecules in the held-out set were sampled from the generative model. b, Chemical structures, left, and sampling frequencies, right, for an illustrative subset of molecules in the held-out set that were correctly anticipated by the generated molecule. The molecules were selected from across the spectrum of sampling frequency in order to illustrate some of the major chemotypes captured by the generative model.
Extended Data Fig. 6 Benchmarking the structural prior using continuous molecular embeddings.
a, Median Euclidean distance between CDDD embeddings of held-out NPSs and generated molecules matching their exact masses ( ± 10 ppm), arranged in descending order by sampling frequency (“most frequent”), ascending order by sampling frequency (“least frequent”), or a random sample of molecules with matching exact masses from PubChem. Error bars show the interquartile range. b, Distribution of Euclidean distances between the CDDD embeddings of held-out NPSs and generated molecules matching their exact masses ( ± 10 ppm), taking either the single most frequently sampled generated molecule, the single least frequently sampled generated molecule, or a random molecule with a matching exact mass from PubChem.
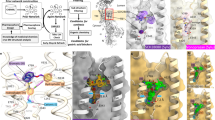
Extended Data Fig. 7 Application of the structural prior to the synthetic cannabinoid ADB-HEXINACA.
Left, the chemical structure, molecular formula, and exact mass of ADB-HEXINACA. Middle, sampling frequencies of the 20 most frequently sampled molecules matching the exact mass of ADB-HEXINACA ( ± a 10 ppm window). An illustrative subset of the generated molecules, highlighted in red, are shown on the bottom.
Extended Data Fig. 8 Improved chemical similarity of automatically elucidated structures after MS/MS data integration.
a, Euclidean distances between CDDD embeddings for molecules in the held-out set of unidentified NPSs and the top-ranked structures suggested by CFM-ID alone, the structural prior alone, or the combination of the two. b, Improvements in automated structure elucidation of an unidentified NPS using tandem mass spectrometry. Left, the chemical structure of 3,4-MDMA methylene homologue, created by inserting a methylene spacer between the α-carbon and amine group in MDMA. Middle, the top-ranked molecule suggested by the structural prior and mirror plot comparing the observed tandem mass spectrum of 3,4-MDMA methylene homologue with the tandem mass spectrum predicted by CFM-ID. Right, the top-ranked molecule after integrating the structural prior with MS/MS evidence (top) and mirror plot comparing the observed and predicted tandem mass spectra.
Extended Data Fig. 9 Evaluation of DarkNPS against two additional baselines.
a–d, Comparison of DarkNPS to chemical database search against known NPSs from the training set. a, Top-1 accuracy with which the complete chemical structures of unidentified NPSs in the held-out set were correctly elucidated by the combination of the structural prior and CFM-ID, or chemical database search against the 1,753 known NPSs in the training set. Searching against the disjoint training set yields a top-1 accuracy of 0%, by definition. b, Top-k accuracy curve for structure elucidation of unidentified NPSs in the held-out set by the combination of the structural prior and CFM-ID, or chemical database search against the 1,753 known NPSs in the training set. Searching against the disjoint training set yields a top-1 accuracy of 0%, by definition. c, Tanimoto coefficients between the held-out set and the top-ranked structures suggested by the combination of the structural prior and CFM-ID, or chemical database search against the 1,753 known NPSs in the training set. Results are not shown for 30 held-out molecules whose masses did not match any molecule in the training set. d, As in c, but showing the Euclidean distances between CDDD embeddings. Results are not shown for 30 held-out molecules whose masses did not match any molecule in the training set. e–h, Comparison of DarkNPS to the AddCarbon model. e, Top-1 accuracy with which the complete chemical structures of unidentified NPSs in the held-out set were correctly elucidated by the combination of the structural prior and CFM-ID, or chemical database search against 34,358 molecules generated by the AddCarbon baseline. f, Top-k accuracy curve for structure elucidation of unidentified NPSs in the held-out set by the combination of the structural prior and CFM-ID, or chemical database search against 34,358 molecules generated by the AddCarbon baseline. g, Tanimoto coefficients between the held-out set and the top-ranked structures suggested by the combination of the structural prior and CFM-ID, or chemical database search against 34,358 molecules generated by the AddCarbon baseline. Results are not shown for 38 held-out molecules whose masses did not match any molecule generated by the AddCarbon model. h, As in g, but showing the Euclidean distances between CDDD embeddings. Results are not shown for 38 held-out molecules whose masses did not match any molecule generated by the AddCarbon model.
Extended Data Fig. 10 Robustness of the structural prior to sample size.
a, Correlation between molecular sampling frequency in the original sample of 1 billion SMILES strings and a second sample of 1 billion SMILES strings. Inset text shows the Pearson correlation. b, Proportion of NPSs in the held-out set that were generated after downsampling the original sample of 1 billion SMILES strings to between 1,000 and 300 million SMILES. Only marginal improvement is observed after approximately 100 million SMILES. c, Top-k accuracy of the structural prior in the held-out set after downsampling the original sample of 1 billion SMILES strings to between 1,000 and 300 million SMILES. Only marginal improvement is observed after approximately 100 million SMILES.
Supplementary information
Rights and permissions
About this article
Cite this article
Skinnider, M.A., Wang, F., Pasin, D. et al. A deep generative model enables automated structure elucidation of novel psychoactive substances. Nat Mach Intell 3, 973–984 (2021). https://doi.org/10.1038/s42256-021-00407-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00407-x
This article is cited by
-
Invalid SMILES are beneficial rather than detrimental to chemical language models
Nature Machine Intelligence (2024)
-
AI is a viable alternative to high throughput screening: a 318-target study
Scientific Reports (2024)
-
67 million natural product-like compound database generated via molecular language processing
Scientific Data (2023)
-
MSNovelist: de novo structure generation from mass spectra
Nature Methods (2022)
-
Developments in high-resolution mass spectrometric analyses of new psychoactive substances
Archives of Toxicology (2022)