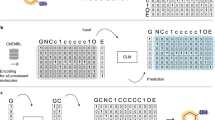
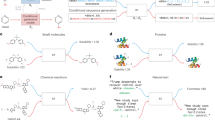
Abstract
Machine learning-based generative models can generate novel molecules with desirable physiochemical and pharmacological properties from scratch. Many excellent generative models have been proposed, but multi-objective optimizations in molecular generative tasks are still quite challenging for most existing models. Here we proposed the multi-constraint molecular generation (MCMG) approach that can satisfy multiple constraints by combining conditional transformer and reinforcement learning algorithms through knowledge distillation. A conditional transformer was used to train a molecular generative model by efficiently learning and incorporating the structure–property relations into a biased generative process. A knowledge distillation model was then employed to reduce the model’s complexity so that it can be efficiently fine-tuned by reinforcement learning and enhance the structural diversity of the generated molecules. As demonstrated by a set of comprehensive benchmarks, MCMG is a highly effective approach to traverse large and complex chemical space in search of novel compounds that satisfy multiple property constraints.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The training dataset was obtained from the study reported by Olivecrona et al.26, which selected the data from the ChEMBL51 dataset. The bioactivity dataset includes the experimental bioactivity data for three different protein targets, namely DRD2, JNK3 and GSK3β. The DRD2 dataset was provided by Olivecrona et al.26, which contains 100,000 negative and 7,219 positive compounds. The JNK3 dataset52 contains the inhibition data for 50,000 negative and 2,665 positive compounds, whereas the GSK3β dataset53,54 contains the inhibition data for 50,000 negative and 740 positive compounds. The JNK3 and GSK3β datasets are available from the study of Li and colleagues41.
Code availability
The code used in the study is publicly available from the GitHub repository: https://github.com/jkwang93/MCMG (ref. 64).
References
Elton, D. C., Boukouvalas, Z., Fuge, M. D. & Chung, P. W. Deep learning for molecular design-a review of the state of the art. Mol. Syst. Design Eng. 4, 828–849 (2019).
Chen, H., Engkvist, O., Wang, Y., Olivecrona, M. & Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 23, 1241–1250 (2018).
Chen, H. & Engkvist, O. Has drug design augmented by artificial intelligence become a reality? Trends Pharmacol. Sci. 40, 806–809 (2019).
Ekins, S. et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019).
Mater, A. C. & Coote, M. L. Deep learning in chemistry. J. Chem. Inf. Model. 59, 2545–2559 (2019).
Jørgensen, P. B., Schmidt, M. N. & Winther, O. Deep generative models for molecular science. Mol. Inf. 37, 1700133 (2018).
Yang, X., Wang, Y., Byrne, R., Schneider, G. & Yang, S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem. Rev. 119, 10520–10594 (2019).
Hessler, G. & Baringhaus, K.-H. Artificial intelligence in drug design. Molecules 23, 2520 (2018).
Batool, M., Ahmad, B. & Choi, S. A structure-based drug discovery paradigm. Int. J. Mol. Sci. 20, 2783 (2019).
Xu, Y. et al. Deep learning for molecular generation. Future Med. Chem. 11, 567–597 (2019).
Button, A., Merk, D., Hiss, J. A. & Schneider, G. Automated de novo molecular design by hybrid machine intelligence and rule-driven chemical synthesis. Nat. Mach. Intell. 1, 307–315 (2019).
Moret, M., Friedrich, L., Grisoni, F., Merk, D. & Schneider, G. Generative molecular design in low data regimes. Nat. Mach. Intell. 2, 171–180 (2020).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Central Sci. 4, 268–276 (2018).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Polykovskiy, D. et al. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharmaceutics 15, 4398–4405 (2018).
Putin, E. et al. Adversarial threshold neural computer for molecular de novo design. Mol. Pharm. 15, 4386–4397 (2018).
Bjerrum, E. J. & Threlfall, R. Molecular generation with recurrent neural networks (RNNs). Preprint at https://arxiv.org/abs/1705.04612 (2017).
Gupta, A. et al. Generative recurrent networks for de novo drug design. Mol. Inf. 37, 1700111 (2018).
Pogány, P., Arad, N., Genway, S. & Pickett, S. D. De novo molecule design by translating from reduced graphs to SMILES. J. Chem. Inf. Model. 59, 1136–1146 (2019).
Liu, X., Ye, K., van Vlijmen, H. W. T., Ijzerman, A. P. & van Westen, G. J. P. An exploration strategy improves the diversity of de novo ligands using deep reinforcement learning: a case for the adenosine A2A receptor. J. Cheminf. 11, 35 (2019).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Central Sci. 4, 120–131 (2018).
Yang, X., Zhang, J., Yoshizoe, K., Terayama, K. & Tsuda, K. ChemTS: an efficient python library for de novo molecular generation. Sci. Technol. Adv. Mater. 18, 972–976 (2017).
Grisoni, F., Moret, M., Lingwood, R. & Schneider, G. Bidirectional molecule generation with recurrent neural networks. J. Chem. Inf. Model. 60, 1175–1183 (2020).
Merk, D., Friedrich, L., Grisoni, F. & Schneider, G. De novo design of bioactive small molecules by artificial intelligence. Mol. Inf. 37, 1700153 (2018).
Popova, M., Isayev, O. & Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 4, eaap7885 (2018).
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 9, 48 (2017).
Lim, J., Ryu, S., Kim, J. W. & Kim, W. Y. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminf. 10, 31 (2018).
Kusner, M. J., Paige, B. & Hernández-Lobato, J. M. in Proc. 34th International Conference on Machine Learning Vol. 70. (eds. Doina, P. & Yee Whye, T.) 1945–1954 (PMLR, 2017).
Liu, Q., Allamanis, M., Brockschmidt, M. & Gaunt, A. L. in Proc. 32nd International Conference on Neural Information Processing Systems 7806–7815 (Curran Associates Inc., 2018).
Simonovsky, M. & Komodakis, N. in International Conference on Artificial Neural Networks 412–422 (Springer, 2018).
Bjerrum, E. J. & Sattarov, B. Improving chemical autoencoder latent space and molecular de novo generation diversity with heteroencoders. Biomolecules 8, 131 (2018).
Jin, W., Barzilay, R. & Jaakkola, T. in Proc. 35th International Conference on Machine Learning Vol. 80. (eds. Jennifer, D. & Andreas, K.) 2323–2332 (PMLR, 2018).
Kang, S. & Cho, K. Conditional molecular design with deep generative models. J. Chem. Inf. Model. 59, 43–52 (2019).
Kingma, D. P. & Welling, M. Auto-encoding variational Bayes. Preprint at https://arxiv.org/abs/1312.6114 (2014).
Kadurin, A., Nikolenko, S., Khrabrov, K., Aliper, A. & Zhavoronkov, A. druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico. Mol. Pharmaceutics 14, 3098–3104 (2017).
Sanchez-Lengeling, B., Outeiral, C., Guimaraes, G. L. & Aspuru-Guzik, A. Optimizing distributions over molecular space. An objective-reinforced generative adversarial network for inverse-design chemistry (ORGANIC). Preprint at ChemRxiv https://doi.org/10.26434/chemrxiv.5309668.v3 (2017).
Guimaraes, G. L., Sanchez-Lengeling, B., Farias, P. L. C. & Aspuru-Guzik, A. Objective-reinforced generative adversarial networks (ORGAN) for sequence generation models. Preprint at https://arxiv.org/abs/1705.10843 (2017).
Putin, E. et al. Reinforced adversarial neural computer for de novo molecular design. J. Chem. Inf. Model. 58, 1194–1204 (2018).
Yu, L., Zhang, W., Wang, J. & Yu, Y. in Proc. 31st AAAI Conference on Artificial Intelligence 2852–2858 (AAAI Press, 2017).
Sohn, K., Yan, X. & Lee, H. in Proc. 28th International Conference on Neural Information Processing Systems Vol. 2, 3483–3491 (MIT Press, 2015).
You, J., Liu, B., Ying, Z., Pande, V. & Leskovec, J. in Advances in Neural Information Processing Systems 6410–6421 (2018).
Brochu, E., Cora, V. M. & Freitas, N. d. A tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. Preprint at https://arxiv.org/abs//1012.2599 (2010).
Cao, N. D. & Kipf, T. MolGAN: an implicit generative model for small molecular graphs. Preprint at https://arxiv.org/abs/1805.11973 (2018).
Jaques, N. et al. in Proc. 34th International Conference on Machine Learning Vol. 70, 1645–1654 (JMLR.org, 2017).
Sutton, R. S. & Barto, A. G. Introduction to Reinforcement Learning (MIT Press, 1998).
Blaschke, T. et al. REINVENT 2.0: an AI tool for de novo drug design. J. Chem. Inf. Model. 60, 5918–5922 (2020).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Proc. Syst. 30, 5998–6008 (2017).
Tripp, A., Daxberger, E. & Hernández-Lobato, J. M. in Advances in Neural Information Processing Systems 11259–11272 (2020).
Bemis, G. W. & Murcko, M. A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Blaschke, T., Engkvist, O., Bajorath, J. & Chen, H. Memory-assisted reinforcement learning for diverse molecular de novo design. J. Cheminf. 12, 68 (2020).
Anna, G. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, 1100–1107 (2012).
Ip, Y. T. & Davis, R. J. Signal transduction by the c-Jun N-terminal kinase (JNK)-from inflammation to development. Curr. Opin. Cell Biol. 10, 205–219 (1998).
Shang, L. et al. RAGE modulates hypoxia/reoxygenation injury in adult murine cardiomyocytes via JNK and GSK-3 beta signaling pathways. PLoS ONE 5, e10092 (2010).
Tanabe, K. et al. Glucose and fatty acids synergize to promote B-cell apoptosis through activation of glycogen synthase kinase 3 beta independent of JNK activation. PLoS ONE 6, e18146 (2011).
Hinton, G., Vinyals, O. & Dean, J. Distilling the knowledge in a neural network. Computer Sci. 14, 38–39 (2015).
Cho, K. et al. Learning phrase representations using RNN Encoder decoder for statistical machine translation. Preprint at https://arxiv.org/abs/1406.1078 (2014).
Jaques, N., Gu, S., Turner, R. E. & Eck, D. Tuning recurrent neural networks with reinforcement learning. Preprint at https://arxiv.org/abs/1611.02796v1 (2017).
Jin, W., Barzilay, R. & Jaakkola, T. Composing molecules with multiple property constraints. Preprint at https://arxiv.org/abs/2002.03244v1 (2020).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
David, R. & Mathew, H. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Freeze, J. G., Kelly, H. R. & Batista, V. S. Search for catalysts by inverse design: artificial intelligence, mountain climbers, and alchemists. Chem. Rev. 119, 6595–6612 (2019).
Polykovskiy, D. et al. Molecular Sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 11, 565644 (2020).
Wang J. et al. Code Repository jkwang93/MCMG: v1.1.0 (Zenodo, 2021); https://doi.org/10.5281/zenodo.5205570
Acknowledgements
We want to thank Z. Liu for insightful discussion on this study. This work was financially supported by National Key R&D Program of China (grant no. 2016YFA0501701), National Natural Science Foundation of China (grant no. 81773632), Natural Science Foundation of Zhejiang Province (grant no. LZ19H300001), Key R&D Program of Zhejiang Province (grant no. 2020C03010), and Fundamental Research Funds for the Central Universities (grant no. 2020QNA7003).
Author information
Authors and Affiliations
Contributions
T.J.H., C.Y.H., D.S.C. and X.C. designed the research study. J.K.W. developed the method and wrote the code. J.K.W., M.Y.W., X.R.W., D.J.J., B.B.L., X.J.Z. B.Y. and Q.J.H. performed the analysis. J.K.W., M.Y.W., T.J.H. and C.Y.H. wrote the paper. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks J.B. Brown, Jose Jimenez-Luna, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Discussion and Tables 1–6.
Rights and permissions
About this article
Cite this article
Wang, J., Hsieh, CY., Wang, M. et al. Multi-constraint molecular generation based on conditional transformer, knowledge distillation and reinforcement learning. Nat Mach Intell 3, 914–922 (2021). https://doi.org/10.1038/s42256-021-00403-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00403-1
This article is cited by
-
PocketFlow is a data-and-knowledge-driven structure-based molecular generative model
Nature Machine Intelligence (2024)
-
Deep generative model for drug design from protein target sequence
Journal of Cheminformatics (2023)
-
DeepSA: a deep-learning driven predictor of compound synthesis accessibility
Journal of Cheminformatics (2023)
-
MolFilterGAN: a progressively augmented generative adversarial network for triaging AI-designed molecules
Journal of Cheminformatics (2023)
-
Bird’s Eye View feature selection for high-dimensional data
Scientific Reports (2023)