Abstract
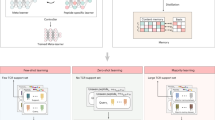
Neoantigens play a key role in the recognition of tumour cells by T cells; however, only a small proportion of neoantigens truly elicit T-cell responses, and few clues exist as to which neoantigens are recognized by which T-cell receptors (TCRs). We built a transfer learning-based model named the pMHC–TCR binding prediction network (pMTnet) to predict TCR binding specificities of the neoantigens—and T cell antigens in general—presented by class I major histocompatibility complexes. pMTnet was comprehensively validated by a series of analyses and exhibited great advances over previous works. By applying pMTnet to human tumour genomics data, we discovered that neoantigens were generally more immunogenic than self-antigens, but human endogenous retrovirus E (a special type of self-antigen that is reactivated in kidney cancer) is more immunogenic than neoantigens. We further discovered that patients with more clonally expanded T cells that exhibit better affinity against truncal rather than subclonal neoantigens had more favourable prognosis and treatment response to immunotherapy in melanoma and lung cancer but not in kidney cancer. Predicting TCR–neoantigen/antigen pairing is one of the most daunting challenges in modern immunology; however, we achieved an accurate prediction of the pairing using only the TCR sequence (CDR3β), antigen sequence and class I major histocompatibility complex allele, and our work revealed unique insights into the interactions between TCRs and major histocompatibility complexes in human tumours, using pMTnet as a discovery tool.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Details on data used for the training and validation of pMTnet, including sample size and role in the machine learning process, are presented in the Supplementary Information. The training and testing datasets are shared on our github repository: https://github.com/tianshilu/pMTnet. The processed TCR-seq and scRNA-seq data generated from the in-house patient donor are also archived at https://github.com/tianshilu/pMTnet. The raw scRNA-seq plus TCR-seq data have been archived on NIH GEO with the accession number GSE173165.
For the NIES analyses, the public patient sequencing datasets are from TCGA, Liu et al.43, Van Allen et al.44 and Hugo et al.45. The raw RNA-seq and exome-seq data of the in-house IL2 cohort patients can be downloaded from the European Genome Phenome Archive with accession number EGAS00001003605 through controlled access. Source data are provided with this paper.
Code availability
The pMTnet software is available on GitHub at https://github.com/tianshilu/pMTnet (ref. 55). Pipeline for HERV expression detection is available on GitHub at https://github.com/jcao89757/HERVranger (ref. 56). QBRC mutation calling pipeline is available on GitHub at https://github.com/tianshilu/QBRC-Somatic-Pipeline (ref. 57). QBRC neoantigen calling pipeline is available on GitHub at https://github.com/tianshilu/QBRC-Neoantigen-Pipeline (ref. 58).
References
Dunn, G. P., Old, L. J. & Schreiber, R. D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 22, 329–360 (2004).
Ascierto, P. A. & Marincola, F. M. 2015: The year of anti-PD-1/PD-L1s against melanoma and beyond. EBioMedicine 2, 92–93 (2015).
Anagnostou, V. et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017).
Reck, M. et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Rizvi, N. A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Linette, G. P. & Carreno, B. M. Neoantigen vaccines pass the immunogenicity test. Trends Mol. Med. 23, 869–871 (2017).
Verdegaal, E. M. E. et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536, 91–95 (2016).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Zhang, S.-Q. et al. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat. Biotechnol. 36, 1156–1159 (2018).
Kula, T. et al. T-Scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178, 1016–1028.e13 (2019).
Ito, A. et al. Cancer neoantigens: a promising source of immunogens for cancer immunotherapy. J. Clin. Cell. Immunol. https://doi.org/10.4172/2155-9899.1000322 (2015).
Hou, X. et al. Analysis of the repertoire features of TCR beta chain CDR3 in human by high-throughput sequencing. Cell. Physiol. Biochem. 39, 651–667 (2016).
Atchley, W. R., Zhao, J., Fernandes, A. D. & Drüke, T. Solving the protein sequence metric problem. Proc. Natl Acad. Sci. USA 102, 6395–6400 (2005).
Zhang, Z., Xiong, D., Wang, X., Liu, H. & Wang, T. Mapping the functional landscape of T cell receptor repertoires by single-T cell transcriptomics. Nat. Methods 18, 92–99 (2021).
Nielsen, M. & Andreatta, M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33 (2016).
Tickotsky, N., Sagiv, T., Prilusky, J., Shifrut, E. & Friedman, N. McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017).
Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017).
Huth, A., Liang, X., Krebs, S., Blum, H. & Moosmann, A. Antigen-specific TCR signatures of cytomegalovirus infection. J. Immunol. 202, 979–990 (2019).
Chen, G. et al. Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens. Cell Rep. 19, 569–583 (2017).
Joglekar, A. V. et al. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods 16, 191–198 (2019).
Bagaev, D. V. et al. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucl. Acids Res. 48, D1057–D1062 (2020).
Zhang, W. et al. PIRD: pan immune repertoire database. Bioinformatics 36, 897–903 (2020).
Jokinen, E., Heinonen, M., Huuhtanen, J., Mustjoki, S. & Lähdesmäki, H. TCRGP: determining epitope specificity of T cell receptors. Preprint at https://www.biorxiv.org/content/10.1101/542332v1 (2019).
Jurtz, V. I. et al. NetTCR: sequence-based prediction of TCR binding to peptide–MHC complexes using convolutional neural networks. Preprint at https://www.biorxiv.org/content/10.1101/433706v1 (2018).
Gielis, S. et al. Detection of enriched T cell epitope specificity in full T cell receptor sequence repertoires. Front. Immunol. 10, 2820 (2019).
Gee, M. H. et al. Antigen identification for orphan T Cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563.e16 (2018).
Liu, Y. C. et al. Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule. J. Biol. Chem. 288, 15442–15454 (2013).
Cole, D. K. et al. T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR–major histocompatibility complex interactions. J. Biol. Chem. 289, 628–638 (2014).
Tran, E. et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015).
Weiss, G. A., Watanabe, C. K., Zhong, A., Goddard, A. & Sidhu, S. S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl Acad. Sci. USA 97, 8950–8954 (2000).
Valkenburg, S. A. et al. Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses. Proc. Natl Acad. Sci. USA 113, 4440–4445 (2016).
Wang, T. et al. An empirical approach leveraging tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors. Cancer Discov. 8, 1142–1155 (2018).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015).
Lo, A. S.-Y., Xu, C., Murakami, A. & Marasco, W. A. Regression of established renal cell carcinoma in nude mice using lentivirus-transduced human T cells expressing a human anti-CAIX chimeric antigen receptor. Mol. Ther. Oncolytics 1, 14003 (2014).
Cherkasova, E. et al. Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res. 76, 2177–2185 (2016).
Reuben, A. et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 7, 1088–1097 (2017).
Lu, T. et al. Tumor neoantigenicity assessment with CSiN score incorporates clonality and immunogenicity to predict immunotherapy outcomes. Sci. Immunol. 5, eaaz3199 (2020).
Simnica, D. et al. T cell receptor next-generation sequencing reveals cancer-associated repertoire metrics and reconstitution after chemotherapy in patients with hematological and solid tumors. Oncoimmunology 8, e1644110 (2019).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016).
Kim, S. T. et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 24, 1449–1458 (2018).
Miao, D. et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018).
Dash, P. et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Nielsen, M. et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS ONE 2, e796 (2007).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Miao, D. et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018).
tianshilu/pMTnet: First Release (Zenodo, 2021); https://doi.org/10.5281/zenodo.4670312
jcao89757/HERVranger: HERVranger (Zenodo, 2021); https://doi.org/10.5281/zenodo.4681560
tianshilu/QBRC-Somatic-Pipeline: First Release (Zenodo, 2021); https://doi.org/10.5281/zenodo.4670314
tianshilu/QBRC-Neoantigen-Pipeline: First Release (Zenodo, 2021); https://doi.org/10.5281/zenodo.4670320
Acknowledgements
The Genotype-Tissue Expression (GTEx) project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 10/01/19. We acknowledge D. Liu, B. Li and J. Ostmeyer from UT Southwestern for their helpful advice on our project. We acknowledge the authors of the phs000452.v3.p153 and phs001493.v1.p154 datasets, as well as the funding agencies that supported these studies and dbGaP that supported the archiving of these datasets. This study was supported by the National Institutes of Health (NIH) (grant nos. CCSG 5P30CA142543/TW and R01CA258584/TW), Cancer Prevention Research Institute of Texas (grant no. CPRIT RP190208/TW), University of Texas MD Anderson Cancer Center (Lung Cancer Moon Shot/AR), the University Cancer Foundation at the University of Texas MD Anderson Cancer Center (Institutional Research Grant/AR), the Waun Ki Hong Lung Cancer Research Fund (A.R.), Exon 20 Group (A.R.) and Rexanna’s Foundation for Fighting Lung Cancer (A.R.).
Author information
Authors and Affiliations
Contributions
T.L. created the TCR/pMHC pairing prediction model and carried out the primary data analyses. Z.Z. created the TCR embedding algorithm. J.Z. carried out structural analyses. P.J., C.B., J.V.H., D.L.G. and A.R. contributed the in-house validation data. A.R., Y.W., X.X., J.W. and L.X. provided input on the study design and reviewed the manuscript. T.W. supervised the whole study.
Corresponding authors
Ethics declarations
Competing interests
We are applying for formal intellectual property protection on the pMTnet model and software. A.R. serves on the Scientific Advisory Board and has received honoraria from Adaptive Biotechnologies.
Additional information
Peer review information Nature Machine Intelligence thanks Alok Joglekar, Peng Jiang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 More examples showing the successful embedding of TCRs by the auto-encoder.
(a) Heatmaps of the original TCR CDR3β sequences, embedded by the ‘Atchley factors’ and all padded with zeros to the length of 80 amino acids. (b) Heatmaps of the re-constructed TCR CDR3β sequences for the same TCRs. (c) Scatterplots showing the consistency between ‘Atchley factor’ values of the original and re-constructed TCRs. Blue points represent tiles in the heatmaps in (a) and (b). The red dashed lines are for y = x.
Extended Data Fig. 2 Differential analysis of the expression levels of HERVs between tumor samples and normal samples in different RCC cancer types and data cohorts.
In addition to EU137846.2 (the known HERV-E), the HERVs whose tumor-over-normal expression ratio is >3 in any of the type/cohort, and whose normal tissue expression is <3 are also shown. There are five such HERVs.
Extended Data Fig. 3 Efficiencies of TCR-neoantigen interactions impact response to immunotherapies.
(a) Association between NIES and overall survival of melanoma patients on immunotherapies. The patients were split by the median of NIES in each cohort and then combined. The P-value for the log-rank test is shown. (b) Association between NIES and the response of metastatic gastric cancer patients. The overall survival or progression-free survival data are not made available from the original publication, so we used the RECIST response variables. Complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). There are 40 gastric cancer patients. An ordinal Jonckheere test is employed to investigate whether patients with better response to immunotherapies also have higher NIES scores. In this test, all categories are compared together to investigate whether an overall trend exists across all categories. (c) Boxplots of bootstrap P values evaluating the robustness of comparison between NIES, neoantigen load, T cell infiltration level, and TCR diversity. One P-value is generated from one bootstrap resample of each cohort, and the two-sided Wilcoxon signed-rank test was carried out for the bootstrap P values to assess whether differences are significant between different biomarkers. NS: P>0.01, *: P = 0.01–0.05, **: P = 0.001–0.01, ***: P = 0.0001–0.001, ****:P < 0.0001. For boxplots in (b) and (c), box boundaries represent interquartile ranges, whiskers extend to the most extreme data point which is no more than 1.5 times the interquartile range, and the line in the middle of the box represents the median.
Extended Data Fig. 4
Association of NIES with treatment response of (a) melanoma, (b) metastatic gastric cancer, and (c) kidney cancer patients on checkpoint-inhibitor treatment. There are 33 kidney cancer patients from the Miao cohort. The same analyses as in Extended Data Fig. 3 were carried out, except that the binding affinity cutoffs for assigning TCRs to neoantigens were varied at several possible values.
Extended Data Fig. 5
Association of neoantigen load, T cell infiltration level, and TCR repertoire diversity with treatment response of (a) melanoma, (b) metastatic gastric cancer, and (c) kidney cancer patients on checkpoint-inhibitor treatment. The same analyses as in Extended Data Fig. 3 were carried out for these biomarkers.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Supplementary Tables 1–5.
Source data
Source Data Fig. 2
Statistical source data for Fig. 2
Source Data Fig. 3
Statistical source data for Fig. 3
Source Data Fig. 4
Statistical source data for Fig. 4
Source Data Fig. 5
Statistical source data for Fig. 5
Source Data Fig. 6
Statistical source data for Fig. 6
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5
Rights and permissions
About this article
Cite this article
Lu, T., Zhang, Z., Zhu, J. et al. Deep learning-based prediction of the T cell receptor–antigen binding specificity. Nat Mach Intell 3, 864–875 (2021). https://doi.org/10.1038/s42256-021-00383-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00383-2
This article is cited by
-
Novel insights into TCR-T cell therapy in solid neoplasms: optimizing adoptive immunotherapy
Experimental Hematology & Oncology (2024)
-
Single-cell immune repertoire analysis
Nature Methods (2024)
-
Deep learning predictions of TCR-epitope interactions reveal epitope-specific chains in dual alpha T cells
Nature Communications (2024)
-
Informing immunotherapy with multi-omics driven machine learning
npj Digital Medicine (2024)
-
Classification of tropical cyclone rain patterns using convolutional autoencoder
Scientific Reports (2024)