Abstract
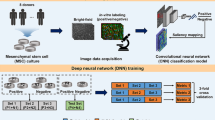
Monoclonalization refers to the isolation and expansion of a single cell derived from a cultured population. This is a valuable step in cell culture that serves to minimize a cell line’s technical variability downstream of cell-altering events, such as reprogramming or gene editing, as well as for processes such as monoclonal antibody development. However, traditional methods for verifying clonality do not scale well, posing a critical obstacle to studies involving large cohorts. Without automated, standardized methods for assessing clonality post hoc, methods involving monoclonalization cannot be reliably upscaled without exacerbating the technical variability of cell lines. Here, we report the design of a deep learning workflow that automatically detects colony presence and identifies clonality from cellular imaging. The workflow, termed Monoqlo, integrates multiple convolutional neural networks and, critically, leverages the chronological directionality of the cell-culturing process. Our algorithm design provides a fully scalable, highly interpretable framework that is capable of analysing industrial data volumes in under an hour using commodity hardware. We focus here on monoclonalization of human induced pluripotent stem cells, but our method is generalizable. Monoqlo standardizes the monoclonalization process, enabling colony selection protocols to be infinitely upscaled while minimizing technical variability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All images from DMR0001, the full monoclonalization run used in the validation of Monoqlo during this study, are available for download from https://nyscf.org/open-source/monoqlo/.
Code availability
The Python code base for executing the Monoqlo framework is available for download at www.nyscf.org/open-source/monoqlo/. For a direct link to the code only, see https://github.com/NYSCF/monoqlo_release (https://zenodo.org/record/4673611).
References
Kwakkenbos, M. J. et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 16, 123–128 (2010).
Wang, G. et al. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat. Protoc. 12, 88–103 (2017).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function and translation. Am. J. Human Genet. 101, 5–22 (2017).
Seki, T., Yuasa, S. & Fukuda, K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat. Protoc. 7, 718–728 (2012).
Chen, Y. H. & Pruett-Miller, S. M. Improving single-cell cloning workflow for gene editing in human pluripotent stem cells. Stem Cell Res. 31, 186–192 (2018).
Paull, D. et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods 12, 885–892 (2015).
Hsieh, C. C. et al. Screening method for rapid identification of hybridomas. US patent 9,797,838 (2017).
Ellis, J. et al. Alternative induced pluripotent stem cell characterization criteria for in vitro applications. Cell Stem Cell 4, 198–199 (2009).
Waisman, A. et al. Deep learning neural networks highly predict very early onset of pluripotent stem cell differentiation. Stem Cell Rep. 12, 845–859 (2019).
Kyttälä, A. et al. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. 6, 200–212 (2016).
Miller, J. D. et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705 (2013).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 1097–1105 (NIPS, 2012).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Wainberg, M., Merico, D., Delong, A. & Frey, B. J. Deep learning in biomedicine. Nat. Biotechnol. 36, 829–838 (2018).
Caicedo, J. C., McQuin, C., Goodman, A., Singh, S. & Carpenter, A. E. Weakly supervised learning of single-cell feature embeddings. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 9309–9318 (IEEE, 2018).
Kusumoto, D. et al. Automated deep learning-based system to identify endothelial cells derived from induced pluripotent stem cells. Stem Cell Rep. 10, 1687–1695 (2018).
Schaub, N. J. et al. Deep learning predicts function of live retinal pigment epithelium from quantitative microscopy. J. Clin. Invest. 130, 1010–1023 (2019).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Pereira, S., Pinto, A., Alves, V. & Silva, C. A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 35, 1240–1251 (2016).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Havaei, M. et al. Brain tumor segmentation with deep neural networks. Med. Image Anal. 35, 18–31 (2017).
Caicedo, J. C. et al. Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat. Methods 16, 1247–1253 (2019).
Girshick, R. Fast R-CNN. In Proc. IEEE International Conference on Computer Vision 1440–1448 (IEEE, 2015).
Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You only look once: unified, real-time object detection. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 779–788 (IEEE, 2016).
Oquab, M., Bottou, L., Laptev, I. & Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 1717–1724 (IEEE, 2014).
Lin, T. Y., Goyal, P., Girshick, R., He, K. & Dollár, P. 2017. Focal loss for dense object detection. In Proc. IEEE International Conference on Computer Vision 2980–2988 (IEEE, 2017).
Agu, C. A. et al. Successful generation of human induced pluripotent stem cell lines from blood samples held at room temperature for up to 48 hr. Stem Cell Rep. 5, 660–671 (2015).
Garg, P. et al. Genome editing of induced pluripotent stem cells to decipher cardiac channelopathy variant. J. Am. College Cardiol. 72, 62–75 (2018).
Rahman, S. H. et al. Rescue of DNA-PK signaling and T-cell differentiation by targeted genome editing in a prkdc deficient iPSC disease model. PLoS Genet. 11, e1005239 (2015).
Warren, C. R. et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell 20, 547–557 (2017).
Wang, Y. et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J. Am. College Cardiol. 64, 451–459 (2014).
Chu, V. T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Smurnyy, Y. et al. DNA sequencing and CRISPR–Cas9 gene editing for target validation in mammalian cells. Nat. Chem. Biol. 10, 623–625 (2014).
Khazaeli, M. B., Conry, R. M. & LoBuglio, A. F. Human immune response to monoclonal antibodies. J. Immunother. Emphasis Tumor Immunol. 15, 42–52 (1994).
Vojtěšek, B., Bartek, J., Midgley, C. A. & Lane, D. P. An immunochemical analysis of the human nuclear phosphoprotein p53: new monoclonal antibodies and epitope mapping using recombinant p53. J. Immunol. Methods 151, 237–244 (1992).
Chen, J. & Srinivas, C. Automatic lymphocyte detection in H&E images with deep neural networks. Preprint at https://arxiv.org/pdf/1612.03217.pdf (2016).
Zhao, J., Zhang, M., Zhou, Z., Chu, J. & Cao, F. Automatic detection and classification of leukocytes using convolutional neural networks. Med. Biol. Eng. Comput. 55, 1287–1301 (2017).
Reichert, J. M. & Valge-Archer, V. E. Development trends for monoclonal antibody cancer therapeutics. Nat. Rev. Drug Discov. 6, 349–356 (2007).
Nelson, A. L., Dhimolea, E. & Reichert, J. M. Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discov. 9, 767–774 (2010).
Christiansen, E. M. et al. In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173, 792–803 (2018).
Kavitha, M. S. et al. Deep vector-based convolutional neural network approach for automatic recognition of colonies of induced pluripotent stem cells. PLoS ONE 12, e0189974 (2017).
Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Perez, L. & Wang, J. The effectiveness of data augmentation in image classification using deep learning. Preprint at https://arxiv.org/pdf/1712.04621.pdf (2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Lin, T. Y. et al. Feature pyramid networks for object detection. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2117–2125 (IEEE, 2017).
Dahl, G. E., Sainath, T. N. & Hinton, G. E. Improving deep neural networks for LVCSR using rectified linear units and dropout. In Proc. 2013 IEEE International Conference on Acoustics, Speech and Signal Processing 8609–8613 (IEEE, 2013).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Preprint at https://arxiv.org/pdf/1502.03167.pdf (2015).
Ren, S., He, K., Girshick, R. & Sun, J. Faster R-CNN: towards real-time object detection with region proposal networks. In Advances in Neural Information Processing Systems 91–99 (NIPS, 2015).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/pdf/1412.6980.pdf (2014).
Jeong, H. J., Park, K. S. & Ha, Y. G. Image preprocessing for efficient training of YOLO deep learning networks. In Proc. 2018 IEEE International Conference on Big Data and Smart Computing (BigComp) 635–637 (IEEE, 2018).
Acknowledgements
This work was supported by The New York Stem Cell Foundation (NYSCF). We thank the members of the NYSCF leadership team, specifically R. Monsma, S. Noggle, R. Aiyar, C. Anzel, L. Schwarzbach, J. Wallerstein and S. Solomon, for their support throughout this work. We also thank L. Mehran and M. Berliss for their guidance on reporting of biological research protocols. We thank C. Richardson for his hugely helpful guidance on the release of Monoqlo.
Author information
Authors and Affiliations
Consortia
Contributions
B.F., D.P. and Z.W. conceptualized the Monoqlo framework, including the use of reverse chronological analysis for the assessment of clonality. B.F. trained and validated RetinaNet detection models and wrote the Python software for the execution and automated deployment of Monoqlo, including data-handling logic, image processing and integration of deep learning models. B.F. conceptualized the use of classification networks in automatically assigning morphological classifications to the most recent colony images. B.F., S.H., B.H., D.P. and J.B. conceptualized the labelling system for classifications of colony morphology. S.H. labelled training data and trained and validated all morphology classification models. G.L. and D.P. developed NYSCF’s iPSC monoclonalization laboratory-automation and colony-selection protocols. B.F., B.H., J.B., D.P. and NYSCF Global Stem Cell Array Team performed image annotations for training the RetinaNet models. D.H., B.H., M.Z., J.B. and NYSCF Global Stem Cell Array Team performed physical monoclonalizations, validation of the Monoqlo framework and subsequent cell culture and imaging using robotic systems.
Corresponding authors
Ethics declarations
Competing interests
B.F., Z.W. and D.P. are co-inventors on a pending patent regarding an image system and method of use (pub. no. WO2021067797A1). The authors declare no other competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Santiago Miriuka, Lassi Paavolainen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
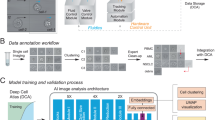
Extended Data Fig. 1 Examples of each morphological class used in training Monoqlo’s classification CNN module.
M1 is the desired morphology, indicating a healthy, pluripotent stem cell colony, and is defined as having a clearly defined, tight perimeter, round shape, no evidence of differentiation and a core with a smooth, transparent appearance. M2 is defined as a colony with the morphology of M1 but with a differentiated fringe. In the displayed example, differentiation and thus loss of pluripotency is clearly shown by the spindle-shaped cell formations and round core with a dark coloration in the bottom left of the tile. M3 is defined as a colony with a poorly-defined shape and often a predominantly dark coloration, which can indicate either differentiation or a dense aggregation of dead cells. M4 is a fully differentiated colony, composed entirely of sprawling, spindle-shaped cell aggregations, and displaying none of the desired morphological markers of pluripotency or iPSC health status.
Extended Data Fig. 2 Example of poor performance by a generalized model trained across all functionalities.
In this instance, the colony detection Is correct. However, the cell detection, in addition to being incorrect, is impossible at the given image magnification and time point.
Extended Data Fig. 3 Predicted Colony Width vs Ground Truth.
Relationship between width of colony bounding box predicted by Monoqlo’s global detection model and the true width measured by biologists with a scale bar image overlay, plotted using 268 measurement-prediction pairs.
Extended Data Fig. 4 Example of abiotic artifacts causing false colony detections by Monoqlo’s global detection model.
a) and b) represent the same image report by Monoqlo, full view and zoomed, respectively.
Extended Data Fig. 5 Example gating strategy.
Representative gating strategy employed during FACS-sort monoclonalization of iPSCs.
Extended Data Fig. 6 Overspill labelling example.
Labelling example in which an additional object class, ‘overspill’ (indicated by blue bounding boxes,) is annotated to improve model performance and mitigate erroneous detections of the ‘colony’ (green bounding box) object class.
Extended Data Fig. 7 Model training and selection.
a, Training and validation accuracy trajectories of the classification CNN, plotted against epoch. Red and green dots signify training and validation accuracies, respectively. b, Confusion matrix of fully trained classification CNN when validated on held-out test set. Scale bar indicates color shading key, indicating the number of examples classified for respective classes as a proportion of total number of examples for the given class. c, Example training and validation accuracy over train time of the RetinaNet detection CNN.
Extended Data Fig. 8 Overlapping detections.
Example of overlapping reports of colonies by Monoqlo’s local detection model where only a single colony exists after ground-truthing.
Extended Data Fig. 9 Colony splitting example.
Illustration of the concept of “colony splitting’, where an apparent single colony is revealed, during reverse-chronological analysis, to have originated in multiple colonies which ultimately merged.
Supplementary information
Supplementary Information
Full details on example neural network architectures from the Monoqlo framework.
Rights and permissions
About this article
Cite this article
Fischbacher, B., Hedaya, S., Hartley, B.J. et al. Modular deep learning enables automated identification of monoclonal cell lines. Nat Mach Intell 3, 632–640 (2021). https://doi.org/10.1038/s42256-021-00354-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00354-7