Abstract
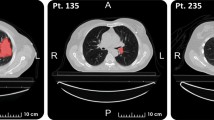
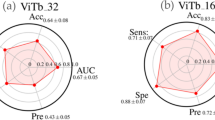
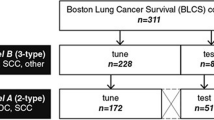
Lung cancer is the most common fatal malignancy in adults worldwide, and non-small-cell lung cancer (NSCLC) accounts for 85% of lung cancer diagnoses. Computed tomography is routinely used in clinical practice to determine lung cancer treatment and assess prognosis. Here, we developed LungNet, a shallow convolutional neural network for predicting outcomes of patients with NSCLC. We trained and evaluated LungNet on four independent cohorts of patients with NSCLC from four medical centres: Stanford Hospital (n = 129), H. Lee Moffitt Cancer Center and Research Institute (n = 185), MAASTRO Clinic (n = 311) and Charité – Universitätsmedizin, Berlin (n = 84). We show that outcomes from LungNet are predictive of overall survival in all four independent survival cohorts as measured by concordance indices of 0.62, 0.62, 0.62 and 0.58 on cohorts 1, 2, 3 and 4, respectively. Furthermore, the survival model can be used, via transfer learning, for classifying benign versus malignant nodules on the Lung Image Database Consortium (n = 1,010), with improved performance (AUC = 0.85) versus training from scratch (AUC = 0.82). LungNet can be used as a non-invasive predictor for prognosis in patients with NSCLC and can facilitate interpretation of computed tomography images for lung cancer stratification and prognostication.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data for cohort 1 (Stanford Hospital, n = 129) are publicly available on The Cancer Imaging Archive (TCIA) at https://doi.org/10.7937/K9/TCIA.2017.7hs46erv (ref. 64). A portion of the data (54/185) for cohort 2 (H. Lee Moffitt Cancer Center and Research Institute, n = 185) is available from TCIA at https://doi.org/10.7937/K9/TCIA.2015.NPGZYZBZ (ref. 65) and https://doi.org/10.7937/K9/TCIA.2015.A6V7JIWX (ref. 66). The data for cohort 3 (MAASTRO Clinic, the Netherlands, n = 311) are publicly available on TCIA at https://doi.org/10.7937/K9/TCIA.2015.PF0M9REI (ref. 11). The data for cohort 4 (Charité – Universitätsmedizin, Berlin, n = 84) are not publicly available yet. The data for LIDC–IDRI (n = 1,010) are available on TCIA at https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX (ref. 67).
Code availability
Code for LungNet is available at https://doi.org/10.24433/CO.0612256.v1 (ref. 68).
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015).
Hirsch, F. R. et al. Lung cancer: current therapies and new targeted treatments. Lancet 389, 299–311 (2017).
Swensen, S. J. et al. CT screening for lung cancer: five-year prospective experience. Radiology 235, 259–265 (2005).
Swensen, S. J. et al. Lung cancer screening with CT: Mayo Clinic experience. Radiology 226, 756–761 (2003).
McWilliams, A. et al. Probability of cancer in pulmonary nodules detected on first screening CT. N. Engl. J. Med. 369, 910–919 (2013).
Henschke, C. I. et al. Early lung cancer action project: overall design and findings from baseline screening. Lancet 354, 99–105 (1999).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278, 563–577 (2015).
Lambin, P. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762 (2017).
Thawani, R. et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer 115, 34–41 (2018).
Zhou, M. et al. Non–small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology (2017).
Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Shen, C. et al. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl. Oncol. 10, 886–894 (2017).
Mattonen, S. A. et al. [18F] FDG positron emission tomography (PET) tumor and penumbra imaging features predict recurrence in non-small cell lung cancer. Tomography 5, 145–153 (2019).
Napel, S., Mu, W., Jardim-Perassi, B. V., Aerts, H. J. W. L. & Gillies, R. J. Quantitative imaging of cancer in the postgenomic era: radio(geno)mics, deep learning, and habitats. Cancer 124, 4633–4649 (2018).
Minamimoto, R. et al. Prediction of EGFR and KRAS mutation in non-small cell lung cancer using quantitative 18F FDG-PET/CT metrics. Oncotarget 8, 52792–52801 (2017).
Gevaert, O. et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 7, 41674 (2017).
van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104–e107 (2017).
Aerts, H. J. W. L. Data science in radiology: a path forward. Clin. Cancer Res. 24, 532–534 (2018).
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H. & Aerts, H. J. W. L. Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510 (2018).
Dehmeshki, J., Amin, H., Valdivieso, M. & Ye, X. Segmentation of pulmonary nodules in thoracic CT scans: a region growing approach. IEEE Trans. Med. Imaging 27, 467–80 (2008).
Lee, Y., Hara, T., Fujita, H., Itoh, S. & Ishigaki, T. Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. IEEE Trans. Med. Imaging 20, 595–604 (2001).
Shen, W., Zhou, M., Yang, F., Yang, C. & Tian, J. in Information Processing in Medical Imaging (eds Ourselin S. et al.) (Springer, 2015).
Xu, Y. et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-18-2495 (2019).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. In Commun. ACM 60, 84–90 (2017).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Bi, W. L. et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J. Clin. 69, 127–157 (2019).
Shin, H. C. et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35, 1285–1298 (2016).
Ardila, D. et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 25, 954–961 (2019).
National Lung Screening Trial Research Team et al. The National Lung Screening Trial: overview and study design. Radiology 258, 243–253 (2011).
National Lung Screening Trial Research Team et al. Results of initial low-dose computed tomographic screening for lung cancer. N. Engl. J. Med. 368, 1980–1991 (2013).
Hanley, J. A. & McNeil, B. J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148, 839–843 (2014).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Jamal-Hanjani, M. et al. Tracking the evolution of non–small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Parmar, C., Grossmann, P., Bussink, J., Lambin, P. & Aerts, H. J. W. L. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 5, 13087 (2015).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. In Proc. 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2009).
Szegedy, C. et al. Going deeper with convolutions. In Proc. 2015 IEEE Conference on Computer Vision and Pattern Recognition 1–9 (IEEE, 2015).
He, K., Zhang, X., Ren S. & Sun, J. Deep residual learning for image recognition. In IEEE Conf. Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Szegedy, C., Ioffe, S., Vanhoucke, V. & Alemi, A. A. Inception-v4, inception-ResNet and the impact of residual connections on learning. In Proc. 31st AAAI Conference on Artificial Intelligence 4278–4284 (AAAI, 2017).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proc. 2017 IEEE Conference on Computer Vision and Pattern Recognition 2261–2269 (IEEE, 2017).
Hara, K., Kataoka, H. & Satoh, Y. Can spatiotemporal 3D CNNs retrace the history of 2D CNNs and ImageNet? In Proc. 2018 IEEE Conference on Computer Vision and Pattern Recognition 6546–6555 (IEEE, 2018).
Raghu, M., Zhang, C., Kleinberg, J. & Bengio, S. Transfusion: understanding Transfer learning for medical imaging. In Advances in Neural Information Processing Systems Vol. 32 (eds H. Wallach et al.) (Curran Associates, 2019).
Causey, J. L. et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci. Rep. 8, 9286 (2018).
Wang, S. et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med. Image Anal. 40, 172–183 (2017).
Zhu, W., Liu, C., Fan, W. & Xie, X. DeepLung: deep 3D dual path nets for automated pulmonary nodule detection and classification. In Proc. 2018 IEEE Winter Conference on Applications of Computer Vision 673–681 (IEEE, 2018).
Shen, W. et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 61, 663–673 (2017).
Cao, H. et al. Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 86, 105934 (2020).
Liu, H. et al. A cascaded dual-pathway residual network for lung nodule segmentation in CT images. Phys. Med. 63, 112–121 (2019).
Hosny, A. et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 15, e1002711 (2018).
Gentles, A. J. et al. Integrating tumor and stromal gene expression signatures with clinical indices for survival stratification of early-stage non-small cell lung cancer. J. Natl Cancer Inst. 107, djv211 (2015).
Liang, C. et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non—small cell lung cancer. Radiology 281, 947–957 (2016).
Shedden, K. et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 14, 822–827 (2008).
Guo, N. L. et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin. Cancer Res. 14, 8213–8220 (2008).
Armato, S. G. et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference: database of lung nodules on CT scans. Med. Phys. 38, 915–931 (2011).
Cox, D. R. Regression models and life-tables. J. R. Stat. Soc. Ser. B 34, 187–220 (1972).
De Boer, P. T., Kroese, D. P., Mannor, S. & Rubinstein, R. Y. A tutorial on the cross-entropy method. Ann. Oper. Res. 134, 19–67 (2005).
Smith, L. N. Cyclical learning rates for training neural networks. In Proc. 2017 IEEE Winter Conference on Applications of Computer Vision 464–472 (IEEE, 2017).
Abadi, M. et al. TensorFlow: a system for large-scale machine learning. Proc. 12th USENIX Conference on Operating Systems Design and Implementation 265–283 (USENIX, 2016).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012).
Gevaert, O. et al. Non–small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data—methods and preliminary results. Radiology 264, 387–396 (2012).
Gevaert, O. et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273, 168–174 (2015).
Huang, C. et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine 45, 70–80 (2019).
Goeman, J. J. L1 penalized estimation in the Cox proportional hazards model. Biom. J. 2, 70–84 (2010).
Davidson-Pilon, C. et al. CamDavidsonPilon/lifelines v0.21.1 (Zenodo, 2019); https://doi.org/10.5281/ZENODO.2652543
Bakr, S. et al. Data descriptor: a radiogenomic dataset of non-small cell lung cancer. Sci. Data 5, 180202 (2018).
Kalpathy-Cramer, J. et al. A comparison of lung nodule segmentation algorithms: methods and results from a multi-institutional study. J. Digit. Imaging 29, 476–487 (2016).
Grove, O. et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS ONE 10, e0118261 (2015).
Armato, S. G. et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung NODULES on CT scans. Med. Phys. 38, 915–931 (2011).
Mukherjee, P., Zhou, M., Lee, E. & Gevaert, O. LungNet: a shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional CT-image data. Code Ocean https://codeocean.com/capsule/5978670/tree/v1 (2020).
Acknowledgements
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number R01EB020527 and R56EB020527. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A Titan X Pascal used for this research was donated by the NVIDIA Corporation.
Author information
Authors and Affiliations
Contributions
Conception design: M.Z., E.L. and O.G. Provision of data: O.G., Y.B., S.N. and R.G. Data analysis and interpretation: P.M., M.Z., E.L. and O.G. Writing: all authors. Computation resource: O.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary information on the features extracted for radiomic analysis.
Rights and permissions
About this article
Cite this article
Mukherjee, P., Zhou, M., Lee, E. et al. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat Mach Intell 2, 274–282 (2020). https://doi.org/10.1038/s42256-020-0173-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-0173-6
This article is cited by
-
Convolutional neural network applied to preoperative venous-phase CT images predicts risk category in patients with gastric gastrointestinal stromal tumors
BMC Cancer (2024)
-
Foundation model for cancer imaging biomarkers
Nature Machine Intelligence (2024)
-
DCCAFN: deep convolution cascade attention fusion network based on imaging genomics for prediction survival analysis of lung cancer
Complex & Intelligent Systems (2024)
-
Classification of lung cancer with deep learning Res-U-Net and molecular imaging
Signal, Image and Video Processing (2024)
-
Mining multi-center heterogeneous medical data with distributed synthetic learning
Nature Communications (2023)