Abstract
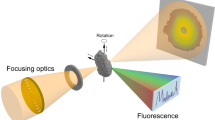
Energy-dispersive X-ray spectroscopy (EDX) is often performed simultaneously with high-angle annular dark-field scanning transmission electron microscopy (STEM) for nanoscale physico-chemical analysis. However, high-quality STEM-EDX tomographic imaging is still challenging due to fundamental limitations such as sample degradation with prolonged scan time and the low probability of X-ray generation. To address this, we propose an unsupervised deep learning method for high-quality 3D EDX tomography of core–shell nanocrystals, which can be usually permanently dammaged by prolonged electron beam. The proposed deep learning STEM-EDX tomography method was used to accurately reconstruct Au nanoparticles and InP/ZnSe/ZnS core–shell quantum dots, used in commercial display devices. Furthermore, the shape and thickness uniformity of the reconstructed ZnSe/ZnS shell closely correlates with optical properties of the quantum dots, such as quantum efficiency and chemical stability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data acquired before and after processing, used to make figures in this paper are publicly shared in the same repository where our code is uploaded (https://github.com/bispl-kaist/Deep-Learning-STEM-EDX-Tomography/ and https://zenodo.org/record/4294003#.X8FJkBNKjOS) under the directory ‘Deep_Learning_STEM-EDX_Tomography/Recon_data’.
Code availability
The codes used in this study are available from https://github.com/bispl-kaist/Deep-Learning-STEM-EDX-Tomography/ and https://zenodo.org/record/4294003#.X8FJkBNKjOS.
References
Alivisatos, A. P. Semiconductor clusters, nanocrystals, and quantum dots. Science 271, 933–937 (1996).
Loss, D. & DiVincenzo, D. P. Quantum computation with quantum dots. Phys. Rev. A 57, 120 (1998).
Won, Y.-H. et al. Highly efficient and stable inp/znse/zns quantum dot light-emitting diodes. Nature 575, 634–638 (2019).
Goldstein, J. I. et al. Scanning Electron Microscopy and X-ray Microanalysis (Springer, 2017).
Lepinay, K., Lorut, F., Pantel, R. & Epicier, T. Chemical 3D tomography of 28 nm high K metal gate transistor: STEM XEDS experimental method and results. Micron 47, 43–49 (2013).
Slater, T. J. et al. STEM-EDX tomography of bimetallic nanoparticles: a methodological investigation. Ultramicroscopy 162, 61–73 (2016).
Saghi, Z., Xu, X., Peng, Y., Inkson, B. & Möbus, G. Three-dimensional chemical analysis of tungsten probes by energy dispersive x-ray nanotomography. Appl. Phys. Lett. 91, 251906 (2007).
Zhong, Z., Goris, B., Schoenmakers, R., Bals, S. & Batenburg, K. J. A bimodal tomographic reconstruction technique combining EDS-STEM and HAADF-STEM. Ultramicroscopy 174, 35–45 (2017).
Gilbert, P. Iterative methods for the three-dimensional reconstruction of an object from projections. J. Theor. Biol. 36, 105–117 (1972).
Kang, E., Min, J. & Ye, J. C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 44, e360–e375 (2017).
Chen, H. et al. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans. Med. Imaging 36, 2524–2535 (2017).
Kang, E., Chang, W., Yoo, J. & Ye, J. C. Deep convolutional framelet denosing for low-dose CT via wavelet residual network. IEEE Trans. Med. Imaging 37, 1358–1369 (2018).
Jin, K. H., McCann, M. T., Froustey, E. & Unser, M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans. Image Process. 26, 4509–4522 (2017).
Han, Y. & Ye, J. C. Framing U-Net via deep convolutional framelets: application to sparse-view CT. IEEE Trans. Med. Imaging 37, 1418–1429 (2018).
Han, Y., Kang, J. & Ye, J. C. Deep learning reconstruction for 9-view dual energy CT baggage scanner. In Proc. Fifth International Conference on Image Formation in X-Ray Computed Tomography 407–410 (CT Meeting, 2018).
Wang, S. et al. Accelerating magnetic resonance imaging via deep learning. In 2016 IEEE 13th International Symposium on Biomedical Imaging 514–517 (IEEE, 2016)..
Hammernik, K. et al. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 79, 3055–3071 (2018).
Lee, D., Yoo, J., Tak, S. & Ye, J. Deep residual learning for accelerated MRI using magnitude and phase networks. IEEE Trans. Biomed. Eng. 65, 1985–1995 (2018).
Han, Y. S., Yoo, J. & Ye, J. C. Deep learning with domain adaptation for accelerated projection reconstruction MR. Magn. Reson. Med. 80, 1189–1205 (2018).
Gong, K. et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans. Med. Imaging 38, 675–685 (2018).
Gong, K., Catana, C., Qi, J. & Li, Q. PET image reconstruction using deep image prior. IEEE Trans. Med. Imaging 38, 1655–1665 (2018).
Luchies, A. C. & Byram, B. C. Deep neural networks for ultrasound beamforming. IEEE Trans. Med. Imaging 37, 2010–2021 (2018).
Yoon, Y. H., Khan, S., Huh, J. & Ye, J. C. Efficient B-mode ultrasound image reconstruction from sub-sampled RF data using deep learning. IEEE Trans. Med. Imaging 38, 325–336 (2019).
Rivenson, Y. et al. Deep learning microscopy. Optica 4, 1437–1443 (2017).
Nehme, E., Weiss, L. E., Michaeli, T. & Shechtman, Y. Deep-STORM: super-resolution single-molecule microscopy by deep learning. Optica 5, 458–464 (2018).
Sinha, A., Lee, J., Li, S. & Barbastathis, G. Lensless computational imaging through deep learning. Optica 4, 1117–1125 (2017).
Donoho, D. L. Compressed sensing. IEEE Trans. Inf. Theory 52, 1289–1306 (2006).
Ye, J. C., Han, Y. & Cha, E. Deep convolutional framelets: a general deep learning framework for inverse problems. SIAM J. Imag. Sci. 11, 991–1048 (2018).
Ye, J. C. & Sung, W. K. Understanding geometry of encoder-decoder CNNs. In Proc. 36th International Conference on Machine Learning (eds Chaudhuri, K. & Salakhutdinov, R.) 7064–7073 (PMLR, 2019)..
Kim, Y. et al. Bright and uniform green light emitting InP/ZnSe/ZnS quantum dots for wide color gamut displays. ACS Appl. Nano Mater. 2, 1496–1504 (2019).
Lee, S. & Lee, C. High-density quantum dots composites and its photolithographic patterning applications. Polym. Adv. Technol. 30, 749–754 (2019).
Virieux, H. et al. InP/ZnS nanocrystals: coupling NMR and XPS for fine surface and interface description. J. Am. Chem. Soc. 134, 19701–19708 (2012).
Stein, J. L. et al. Probing surface defects of InP quantum dots using phosphorus Kα and Kβ X-ray emission spectroscopy. Chem. Mater. 30, 6377–6388 (2018).
Hahm, D. et al. Design principle for bright, robust, and color-pure InP/ZnSexS1–x/ZnS heterostructures. Chem. Mater. 31, 3476–3484 (2019).
Reid, K. R., McBride, J. R., Freymeyer, N. J., Thal, L. B. & Rosenthal, S. J. Chemical structure, ensemble and single-particle spectroscopy of thick-shell inp–znse quantum dots. Nano Lett. 18, 709–716 (2018).
Saxton, W. & Baumeister, W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127, 127–138 (1982).
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention 424–432 (Springer, 2016).
Acknowledgements
This research was supported by Samsung.
Author information
Authors and Affiliations
Contributions
J.C.Y. and E.L. supervised the project in conception and discussion. J.J., E.L. and J.C.Y. co-conceived the project. Y.H., E.C. and H.C. performed experiments for 3D tomography reconstruction. T.-G.K. and S.J. prepared the SAIT QD samples. J.L. performed TEM experiment and image reconstruction using SIRT. M.J., B.G.C. and H.G.K. performed TEM experiments. M.J. and J.J. performed image analysis. S.H. discussed the experiment and co-directed the project. Y.H., J.J., E.C., J.L., H.C., E.L. and J.C.Y. wrote manuscript.
Corresponding authors
Ethics declarations
Competing interests
Y.H., E.C., H.C. and J.C.Y. were supported by a research grant from Samsung Electronics. J.J., J.L., M.J., T.-G.K., B.G.C., H.G.K., S.J., S.H. and E.L. are employees of the Samsung Electronics. The authors declare no other competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 3D reconstruction results of Au-Nanorod for HAADF and EDX measurement.
(a) HAADF ground-truth, and (b) proposed EDX reconstruction.
Extended Data Fig. 2 Comparison of reconstructions using conventional methods and the proposed method.
(a-c) Conventional reconstruction results for SIRT, l1 regularization, and total variation (TV) penalty with spatial 11 × 11 average filter to reduce the measurement noises and (d) our reconstruction results of commercially available QDs. (i) shows the 3D rendering results, and red, green, and blue boxes in (i) indicate (ii) xy- (iii) yz-, and (iv) xz-plane cut-view images, respectively. Yellow box illustrates the magnified images. Se, S, and Zn refer selenium, sulphur, and zinc components, respectively.
Extended Data Fig. 3 Conventional EDX images of sQD1 and sQD2.
(a) sQD1 and (b) sQD2 images acquired using Titan Cubed TEM 60-300 at 300kV, 8 μs dwell time. Se, S, and Zn refer selenium, sulphur, and zinc components, respectively.
Extended Data Fig. 4 Conventional method for analysing QD shape information in TEM images.
sQD images were acquired using Titan Cubed TEM 60-300 at 300 kV, 8 μs dwell time. Total number of particles are 338 and 264 for sQD1 and sQD2, respectively. The image analysis was done with Image J. To isolate particle and background, we calculate mean value of the total images to use as the threshold value. The diameter is calculated as equivalent circular area diameter using the number of counted pixels on each particle.
Extended Data Fig. 5 Photoluminescence characteristics of sQD1 and sQD2.
(a) and (b) are PL-PLE maps of sQD1 and sQD2 dispersed in toluene. Yellow arrows indicate the characteristic points of sQD2 compared sQD1. (c) Absorption and PL spectra for sQD1 and sQD2. (d) Comparison of their quantum efficiency.
Extended Data Fig. 6 SIRT 3D STEM-EDX reconstruction results with spatial 11 × 11 average filter.
(a) and (b) show the reconstruction results using sQD1 and sQD2, respectively. (i) shows the 3D rendering results, and red, green, and blue boxes in (i) indicate (ii) xy-, (iii) yz-, and (iv) xz-plane cut-view images, respectively. Yellow boxes illustrate the magnified images. Se, S, and Zn refer selenium, sulphur, and zinc components, respectively.
Extended Data Fig. 7 sQD1-1 thickness maps on spherical coordinates Φ and θ.
(a-c) sQD1-1 particle 2-D transformed element images; Red, green, and blue represent Se, S, and Zn, respectively. Se, S, and Zn thickness maps at 0∘≤Φ < 360∘ and − 90∘≤θ≤90∘. (d-f) Histogram of Se, S, and Zn thickness maps. Insets are projection images of Se, S, and Zn along z-axis. Colorbars are the number of accumulated pixels at each Φ and θ values.
Extended Data Fig. 8 sQD1-2 thickness maps on spherical coordinate Φ and θ.
(a-c) sQD1-2 particle 2-D transformed element images; Red, green, and blue represent Se, S, and Zn, respectively. Se, S, and Zn thickness maps at 0∘≤Φ < 360∘ and − 90∘≤θ≤90∘. (d-f) Histogram of Se, S, and Zn thickness maps. Insets are projection images of Se, S, and Zn along z-axis. Colorbars are the number of accumulated pixels at each Φ and θ values.
Extended Data Fig. 9 sQD2-1 thickness maps on spherical coordinate Φ and θ.
(a-c) sQD2-1 particle 2-D transformed element images; Red, green, and blue represent Se, S, and Zn, respectively. Se, S, and Zn thickness maps at 0∘≤Φ < 360∘ and − 90∘≤θ≤90∘. (d-f) Histogram of Se, S, and Zn thickness maps. Insets are projection images of Se, S, and Zn along z-axis. Colorbars are the number of accumulated pixels at each Φ and θ values.
Extended Data Fig. 10 sQD2-2 thickness maps on spherical coordinate Φ and θ.
(a-c) sQD2-2 particle 2-D transformed element images; Red, green, and blue represent Se, S, and Zn, respectively. Se, S, and Zn thickness maps at 0∘≤Φ < 360∘ and − 90∘≤θ≤90∘. (d-f) Histogram of Se, S, and Zn thickness maps. Insets are projection images of Se, S, and Zn along z-axis. Colorbars are the number of accumulated pixels at each Φ and θ values.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13, discussion and Table 1.
Rights and permissions
About this article
Cite this article
Han, Y., Jang, J., Cha, E. et al. Deep learning STEM-EDX tomography of nanocrystals. Nat Mach Intell 3, 267–274 (2021). https://doi.org/10.1038/s42256-020-00289-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-00289-5
This article is cited by
-
No ground truth needed: unsupervised sinogram inpainting for nanoparticle electron tomography (UsiNet) to correct missing wedges
npj Computational Materials (2024)
-
A deep learning approach for semantic segmentation of unbalanced data in electron tomography of catalytic materials
Scientific Reports (2022)
-
Identifying chemically similar multiphase nanoprecipitates in compositionally complex non-equilibrium oxides via machine learning
Communications Materials (2022)
-
Five-second STEM dislocation tomography for 300 nm thick specimen assisted by deep-learning-based noise filtering
Scientific Reports (2021)