Abstract
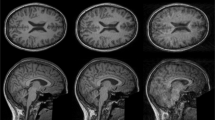
Retrospective artefact correction (RAC) improves image quality post acquisition and enhances image usability. Recent machine-learning-driven techniques for RAC are predominantly based on supervised learning, so practical utility can be limited as data with paired artefact-free and artefact-corrupted images are typically insufficient or even non-existent. Here we show that unwanted image artefacts can be disentangled and removed from an image via an RAC neural network learned with unpaired data. This implies that our method does not require matching artefact-corrupted data to be either collected via acquisition or generated via simulation. Experimental results demonstrate that our method is remarkably effective in removing artefacts and retaining anatomical details in images with different contrasts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data used in this paper were provided by the investigative team of the UNC/UMN Baby Connectome Project. The data can be obtained from the National Institute of Mental Health Data Archive (NDA) (http://nda.nih.gov/) or by contacting the investigative team23.
Code availability
The source code and trained models for this study are publicly available on Zenodo (https://zenodo.org/record/3742351)34.
Change history
27 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s42256-021-00300-7
References
Budde, J., Shajan, G., Scheffler, K. & Pohmann, R. Ultra-high resolution imaging of the human brain using acquisition-weighted imaging at 9.4 T. Neuroimage 86, 592–598 (2014).
Zhuo, J. & Gullapalli, R. P. MR artifacts, safety and quality control. Radiographics 26, 275–297 (2006).
Andre, J. et al. Toward quantifying the prevalence, severity and cost associated with patient motion during clinical MR examinations. J. Am. Coll. Radiol. 12, 689–695 (2015).
Zaitsev, M., Maclaren, J. & Herbst, M. Motion artifacts in MRI: a complex problem with many partial solutions. J. Magn. Reson. Imaging 42, 887–901 (2015).
Zaitsev, M., Dold, C., Sakas, G., Hennig, J. & Speck, O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage 31, 1038–1050 (2006).
Qin, L. et al. Prospective head-movement correction for high-resolution MRI using an in-bore optical tracking system. Magn. Reson. Med. 62, 924–934 (2009).
Ooi, M. B., Krueger, S., Thomas, W. J., Swaminathan, S. V. & Brown, T. R. Prospective real-time correction for arbitrary head motion using active markers. Magn. Reson. Med. 62, 943–954 (2009).
Schulz, J. et al. An embedded optical tracking system for motion-corrected magnetic resonance imaging at 7 T. Magn. Reson. Mater. Phys. Biol. Med. 25, 443–453 (2012).
Maclaren, J. et al. Measurement and correction of microscopic head motion during magnetic resonance imaging of the brain. PLoS ONE 7, e48088 (2012).
Maclaren, J., Herbst, M., Speck, O. & Zaitsev, M. Prospective motion correction in brain imaging: a review. Magn. Reson. Med. 69, 621–636 (2012).
Pipe, J. G. Motion correction with propeller MRI: application to head motion and freebreathing cardiac imaging. Magn. Reson. Med. 42, 963–969 (1999).
Vertinsky, A. T. et al. Performance of PROPELLER relative to standard FSE T2-weighted imaging in pediatric brain MRI. Pediatric Radiol. 39, 1038–1047 (2009).
Jin, K. H., McCann, M. T., Froustey, E. & Unser, M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans. Image Process. 26, 4509–4522 (2017).
Haskell, M. W. et al. Network accelerated motion estimation and reduction (NAMER): convolutional neural network guided retrospective motion correction using a separable motion model. Magn. Reson. Med. 82, 1452–1461 (2019).
Johnson, P. M. & Drangova, M. Motion correction in MRI using deep learning. In Proc. 26th Annual Meeting ISMRM 4098 (ISMRM, 2018).
Tamada, D., Kromrey, M.-L., Ichikawa, S., Onishi, H. & Motosugi, U. Motion artifact reduction using a convolutional neural network for dynamic contrast enhanced MR imaging of the liver. Magn. Reson. Med. Sci. 19, 64–76 (2020).
Küstner, T. et al. Retrospective correction of motion-affected MR images using deep learning frameworks. Magn. Reson. Med. 82, 1527–1540 (2019).
Johnson, P. M. & Drangova, M. Conditional generative adversarial network for 3D rigid-body motion correction in MRI. Magn. Reson. Med. 82, 901–910 (2019).
Liu, M.-Y., Breuel, T. & Kautz, J. Unsupervised image-to-image translation networks. In Proc. Advances in Neural Information Processing Systems 700–708 (NIPS, 2017).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proc. 2017 IEEE International Conference on Computer Vision (ICCV) 2223–2232 (IEEE, 2017).
Zhu, J.-Y. et al. Toward multimodal image-to-image translation. In Proc. Advances in Neural Information Processing Systems 465–476 (NIPS, 2017).
Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In Proc. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 1125–1134 (IEEE, 2017).
Howell, B. R. et al. The UNC/UMN baby connectome project (BCP): an overview of the study design and protocol development. Neuroimage 185, 891–905 (2019).
Perlin, K. An image synthesizer. ACM SIGGRAPH Comput. Graph. 19, 287–296 (1985).
Wang, Z., Bovik, A., Sheikh, H. & Simoncelli, E. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Wang, Z., Simoncelli, E. & Bovik, A. Multiscale structural similarity for image quality assessment. In Proc. 37th IEEE Asilomar Conference on Signals, Systems and Computers 1398–1402 (IEEE, 2003).
Sheikh, H. & Bovik, A. Image information and visual quality. IEEE Trans. Image Process. 15, 430–444 (2006).
Wang, Z. & Bovik, A. A universal image quality index. IEEE Signal Process. Lett. 9, 81–84 (2002).
Mao, X. et al. Least squares generative adversarial networks. In Proc. 2017 IEEE International Conference on Computer Vision (ICCV) 2794–2802 (IEEE, 2017).
Smith, S. M. Fast robust automated brain extraction. Human Brain Mapp. 17, 143–155 (2002).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Ulyanov, D., Vedaldi, A. & Lempitsky, V. Improved texture networks: maximizing quality and diversity in feed-forward stylization and texture synthesis. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 6924–6932 (IEEE, 2017).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proc. International Conference on Learning Representations 1–14 (CLR, 2015).
Liu, S. et al. Code used in “Learning MRI artefact removal with unpaired data”. Zenodo https://doi.org/10.5281/zenodo.37442351 (2020).
Acknowledgements
This work was supported in part by National Institutes of Health grants (EB006733, AG053867, MH117943, MH104324, MH110274) and the efforts of the UNC/UMN Baby Connectome Project Consortium. The authors thank X. Zong of the University of North Carolina at Chapel Hill for an initial discussion on motion artefact simulation and Y. Hong of the University of North Carolina at Chapel Hill and Y. Chen of Case Western Reserve University for proofreading the manuscript.
Author information
Authors and Affiliations
Contributions
S.L. designed the framework and network architecture, carried out the implementation, performed the experiments and analysed the data. S.L. and P.-T.Y. wrote the manuscript. S.L., K.-H.T. and P.-T.Y. revised the manuscript. L.Q. contributed to the initial formulation of the method before moving to Stanford University. W.L. provided the infant data for training and testing. P.-T.Y. conceived the study and were in charge of overall direction and planning. D.S. was involved in the initial discussion of the problem when he was with the University of North Carolina at Chapel Hill. All work was done at the University of North Carolina at Chapel Hill.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Peer review information Nature Machine Intelligence thanks Chuyang Ye and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion and Supplementary Figs. 1–12.
Rights and permissions
About this article
Cite this article
Liu, S., Thung, KH., Qu, L. et al. Learning MRI artefact removal with unpaired data. Nat Mach Intell 3, 60–67 (2021). https://doi.org/10.1038/s42256-020-00270-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-00270-2
This article is cited by
-
Learning multi-site harmonization of magnetic resonance images without traveling human phantoms
Communications Engineering (2024)
-
Stop moving: MR motion correction as an opportunity for artificial intelligence
Magnetic Resonance Materials in Physics, Biology and Medicine (2024)
-
Differential privacy preserved federated transfer learning for multi-institutional 68Ga-PET image artefact detection and disentanglement
European Journal of Nuclear Medicine and Molecular Imaging (2023)
-
Movement-related artefacts (MR-ART) dataset of matched motion-corrupted and clean structural MRI brain scans
Scientific Data (2022)
-
Deep Learning for Image Enhancement and Correction in Magnetic Resonance Imaging—State-of-the-Art and Challenges
Journal of Digital Imaging (2022)