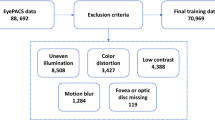
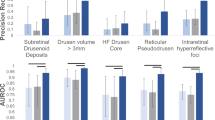
Abstract
Access to large, annotated samples represents a considerable challenge for training accurate deep-learning models in medical imaging. Although at present transfer learning from pre-trained models can help with cases lacking data, this limits design choices and generally results in the use of unnecessarily large models. Here we propose a self-supervised training scheme for obtaining high-quality, pre-trained networks from unlabelled, cross-modal medical imaging data, which will allow the creation of accurate and efficient models. We demonstrate the utility of the scheme by accurately predicting retinal thickness measurements based on optical coherence tomography from simple infrared fundus images. Subsequently, learned representations outperformed advanced classifiers on a separate diabetic retinopathy classification task in a scenario of scarce training data. Our cross-modal, three-stage scheme effectively replaced 26,343 diabetic retinopathy annotations with 1,009 semantic segmentations on optical coherence tomography and reached the same classification accuracy using only 25% of fundus images, without any drawbacks, since optical coherence tomography is not required for predictions. We expect this concept to apply to other multimodal clinical imaging, health records and genomics data, and to corresponding sample-starved learning problems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Since this project uses private patient information the data necessary for reproducing all results cannot be shared. For OCT tissue segmentation, the OCT images and their annotations used for tissue segmentation will be made public and accessible through the public code repository. See thickness_segmentation_data.tar.gz at https://zenodo.org/record/3626020#.X2xYc3ZfjCI44. For the thickness map calculation and prediction, two example DICOM files with OCT volumes together with matching infrared fundus images are provided in the public code repository. This allows simple examples to be carried out and readers can see how the input and output data of the different software packages function. Owing to their data privacy policy, the LMU eye clinic is not able to provide access to the full dataset of 121,985 infrared fundus images and OCT volumes outside of the collaboration between the eye hospital and Helmholtz Zentrum München. Although this inhibits the reproducibility of the thickness prediction and screening evaluation results, the weights from the DeepRT model are shared, enabling full reproducibility of the transfer learning onto the Diabetic Retinopathy dataset. The weights and an example partition of the public diabetic retinopathy detection dataset are made available in the code repository. To reproduce the full study of transfer learning properties of DeepRT compared to ImageNet initialization download the full public Color Fundus Kaggle Diabetic Retinopathy dataset at https://www.kaggle.com/c/diabetic-retinopathy-detection/data. Readers should follow the instructions in the public repository for more detailed steps to reproduce the study. https://www.kaggle.com/c/diabetic-retinopathy-detection/data. The labels for the entire training set and test set are provided in the public code repository. For the evaluation, only the public test dataset is used. For the retinal screening evaluation, none of the patient data can be shared and all publicly available information is in the figures.
Code availability
All trained models as well as all code will be open source, enabling reproducibility of the results from the thickness segmentation and transfer learning on diabetic retinopathy. The code used for the above steps is made available through: https://github.com/theislab/DeepRT45.
References
Ting, D. S. W. et al. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 103, 167–175 (2019).
Schmidt-Erfurth, U., Sadeghipour, A., Gerendas, B. S., Waldstein, S. M. & Bogunović, H. Artificial intelligence in retina. Prog. Retin. Eye Res. 67, 1–29 (2018).
Rajalakshmi, R. et al. Validation of smartphone based retinal photography for diabetic retinopathy screening. PLoS One 10, e0138285 (2015).
Ting, D. S. W. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318, 2211–2223 (2017).
Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016).
De Fauw, J. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 24, 1342–1350 (2018).
Schlegl, T. et al. Fully automated detection and quantification of macular fluid in oct using deep learning. Ophthalmology 125, 549–558 (2018).
Raghu, M., Zhang, C., Kleinberg, J. & Bengio, S. Transfusion: understanding Transfer learning for medical imaging. In Advances in Neural Information Processing Systems 32 (eds. Wallach, H. et al.) 3347–3357 (Curran Associates, 2019).
Bengio, Y. Deep learning of representations for unsupervised and transfer learning. In Proc. International Conference on Machine Learning Workshop on Unsupervised And Transfer Learning 17–36 (ICML, 2012).
Yosinski, J., Clune, J., Bengio, Y. & Lipson, H. How transferable are features in deep neural networks? In Advances in Neural Information Processing Systems Vol. 27 (eds. Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N. D. & Weinberger, K. Q.) 3320–3328 (Curran Associates, 2014).
Deng, J. et al. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conf. on Computer Vision and Pattern Recognition https://doi.org/10.1109/cvprw.2009.5206848 (IEEE, 2009).
Jing, L. & Tian, Y. Self-supervised visual feature learning with deep neural networks: a survey. IEEE Trans. Pattern Anal. Mach. Intell. https://doi.org/10.1109/TPAMI.2020.2992393 (2020).
Pathak, D., Krahenbuhl, P., Donahue, J., Darrell, T. & Efros, A. A. Context encoders: feature learning by inpainting. In 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) https://doi.org/10.1109/cvpr.2016.278 (IEEE, 2016).
Hénaff, O. J., Razavi, A., Doersch, C., Ali Eslami, S. M. & van den Oord, A. Data-efficient image recognition with contrastive predictive coding. Preprint at https://arxiv.org/abs/1905.09272 (2019).
Arandjelovic, R. & Zisserman, A. Look, listen and learn. In Proc. IEEE Int. Conf. on Computer Vision 609–617 (IEEE, 2017).
Sayed, N., Brattoli, B. & Ommer, B. Cross and learn: cross-modal self-supervision. In Pattern Recognition 228–243 (Springer International Publishing, 2019).
Mathis, T. & Kodjikian, L. Five-year outcomes with anti-vascular endothelial growth factor in neovascular age-related macular degeneration: results of the comparison of age-related macular degeneration treatments trials. Ann. Eye Sci. 2, 14 (2018).
Freund, K. B. et al. Type 3 neovascularization: the expanded spectrum of retinal angiomatous proliferation. Retina 28, 201–211 (2008).
Cheung, C. M. G. et al. Improved detection and diagnosis of polypoidal choroidal vasculopathy using a combination of optical coherence tomography and optical coherence tomography angiography. Retina 39, 1655–1663 (2019).
Kortüm, K. U. et al. Using electronic health records to build an ophthalmologic data warehouse and visualize patients’ data. Am. J. Ophthalmol. 178, 84–93 (2017).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. In Lecture Notes in Computer Science 234–241 (Springer, 2015).
Grover, S., Murthy, R. K., Brar, V. S. & Chalam, K. V. Normative data for macular thickness by high-definition spectral-domain optical coherence tomography (spectralis). Am. J. Ophthalmol. 148, 266–271 (2009).
Menke, M. N., Dabov, S., Knecht, P. & Sturm, V. Reproducibility of retinal thickness measurements in patients with age-related macular degeneration using 3D Fourier-domain optical coherence tomography (OCT) (Topcon 3D-OCT 1000). Acta Ophthalmol. 89, 346–351 (2011).
Levandowsky, M. & Winter, D. Distance between sets. Nature 234, 34–35 (1971).
Early Treatment Diabetic Retinopathy Study Research Group. Treatment techniques and clinical guidelines for photocoagulation of diabetic macular edema. Early treatment diabetic retinopathy study report number 2. Ophthalmology 94, 761–774 (1987).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. Preprint at https://arxiv.org/abs/1512.03385 (2015).
Ruamviboonsuk, P. et al. Deep learning versus human graders for classifying diabetic retinopathy severity in a nationwide screening program. npj Digital Medicine 2, 25 (2019).
Lane, N. D. et al. Squeezing deep learning into mobile and embedded devices. IEEE Pervasive Comput. 16, 82–88 (2017).
Ngiam, J. et al. Multimodal deep learning. In Proc. Int. Conf. Machine Learning 689–696 (ICML, 2011).
Baltrusaitis, T., Ahuja, C. & Morency, L.-P. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 41, 423–443 (2019).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proc. IEEE Int. Conf. on Computer Vision 2223–2232 (IEEE, 2017).
Tan, M. & Le, Q. V. EfficientNet: rethinking model scaling for convolutional neural networks. Preprint at https://arxiv.org/abs/1905.11946 (2019).
Huang, D. et al. Optical coherence tomography. Science 254, 1178–1181 (1991).
Barteselli, G. et al. Accuracy of the Heidelberg Spectralis in the alignment between near-infrared image and tomographic scan in a model eye: a multicenter study. Am. J. Ophthalmol. 156, 588–592 (2013).
van Dijk H. W. et al. Selective loss of inner retinal layer thickness in type 1 diabetic patients with minimal diabetic retinopathy. Invest. Opthalm. Visual Sci. 50, 3404–3409 (2009).
Freeman S. R. et al. Optical coherence tomography−raster scanning and manual segmentation in determining drusen volume in age-related macular degeneration. Retina 30, 431–435 (2010).
DeBuc, C. D. & Somfai, G. M. Early detection of retinal thickness changes in diabetes using optical coherence tomography. Med. Sci. Monit. 16, 15–21 (2010).
Arichika S, et. al. Correlation between thickening of the inner and outer retina and visual acuity in patients with epiretinal membrane. Retina 30, 503–508 (2010).
Wada, K. LabelMe: Image Polygonal Annotation with Python https://github.com/wkentaro/labelme (2016).
Horvath, M. M. et al. Modular design, application architecture, and usage of a self-service model for enterprise data delivery: the Duke Enterprise Data Unified Content Explorer (DEDUCE). J. Biomed. Inform. 52, 231–242 (2014).
Golabbakhsh, M. & Rabbani, H. Vessel-based registration of fundus and optical coherence tomography projection images of retina using a quadratic registration model. IET Image Process. 7, 768–776 (2013).
Wu, L., Fernandez-Loaiza, P. & Sauma, J. Classification of diabetic retinopathy and diabetic macular edema. World J. Diabetes 4, 290–294 (2013).
Holmberg, O. et al. Self-supervised retinal thickness prediction enables deep learning from unlabeled data to boost classification of diabetic retinopathy. Zenodo https://doi.org/10.1038/10.5281/zenodo.3626854 (2020).
Holmberg, O. theislab/DeepRT: v0.0.1. Zenodo https://doi.org/10.5281/zenodo.4116029 (2020).
Acknowledgements
This work was supported by the BMBF (grant no. 031L0210A) and by Helmholtz Association’s Initiative and Networking Fund through Helmholtz AI (grant no. ZT-I-PF-5-01). We thank the administrative teams at Helmholtz Zentrum Munich and LMU’s University Hospital (LMU-UH) for the fast data-sharing agreement. We thank LMU-UH data protection officer G. Meyer for constructive and fast approval of data processing. We acknowledge the previous work of M. Müller for setting up and maintaining the data warehouse at LMU-UH and A. Anschütz for its continuation. We thank R. Wolff and A. Babenko for creating the electronic medical records in our hospital and C. Kern for the medical oversight. We thank M. Rohm and I. Manakov for discussions on our data.
Author information
Authors and Affiliations
Contributions
O.G.H. developed the deep learning models and the data analysis pipeline. F.J.T. and N.D.K. conceived the study. K.U.K. led the data acquisition and data interpretation. T.M., J.S., T.H., L.K., B.A. and J.S. performed image annotations and screening evaluations. N.K. supervised the study with F.J.T. and K.U.K. F.J.T., N.D.K., O.G.H. and K.U.K. wrote the paper. O.G.H., N.D.K., K.U.K. and F.J.T. contributed to the interpretation of the results. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
F.J.T. reports receiving consulting fees from Roche Diagnostics GmbH and Cellarity Inc., and ownership interest in Cellarity, Inc. and Dermagnostix. N.D.K. reports ownership interest in Hellsicht GmbH. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 6–15, Supplementary Discussion and Supplementary Table 1–3
Rights and permissions
About this article
Cite this article
Holmberg, O.G., Köhler, N.D., Martins, T. et al. Self-supervised retinal thickness prediction enables deep learning from unlabelled data to boost classification of diabetic retinopathy. Nat Mach Intell 2, 719–726 (2020). https://doi.org/10.1038/s42256-020-00247-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-00247-1
This article is cited by
-
Enhancing pediatric pneumonia diagnosis through masked autoencoders
Scientific Reports (2024)
-
Fast Real-Time Brain Tumor Detection Based on Stimulated Raman Histology and Self-Supervised Deep Learning Model
Journal of Imaging Informatics in Medicine (2024)
-
Automatic diagnosis of keratitis using object localization combined with cost-sensitive deep attention convolutional neural network
Journal of Big Data (2023)
-
Predicting EGFR mutational status from pathology images using a real-world dataset
Scientific Reports (2023)
-
Uncertainty-guided dual-views for semi-supervised volumetric medical image segmentation
Nature Machine Intelligence (2023)