Abstract
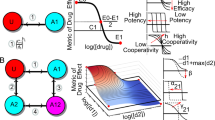
High-throughput drug combination screening provides a systematic strategy to discover unexpected combinatorial synergies in pre-clinical cell models. However, phenotypic combinatorial screening with multi-dose matrix assays is experimentally expensive, especially when the aim is to identify selective combination synergies across a large panel of cell lines or patient samples. Here, we implement DECREASE, an efficient machine learning model that requires only a limited set of pairwise dose–response measurements for accurate prediction of drug combination synergy in a given sample. Using a compendium of 23,595 drug combination matrices tested in various cancer cell lines and malaria and Ebola infection models, we demonstrate how cost-effective experimental designs with DECREASE capture almost the same degree of information for synergy and antagonism detection as the fully measured dose–response matrices. Measuring only the matrix diagonal provides an accurate and practical option for combinatorial screening. The minimal-input web implementation enables applications of DECREASE to both pre-clinical and translational studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The unpublished in-house anticancer dose–response matrix data for all 210 combinations are available either from the DECREASE web tool (http://decrease.fimm.fi/data_availability) or the GitHub repository (https://github.com/IanevskiAleksandr/DECREASE/tree/master/210_Novel_Anticancer_combinations). The published DLBCL anticancer dose–response matrix data are available at the Tripod portal (http://tripod.nih.gov/matrix-data/m3-btk-6x6-ls)11. The data for 22,737 anticancer drug combination from the study of O’Neil and others3 were downloaded from the original paper (http://mct.aacrjournals.org/content/15/6/1155.figures-only#fragments-additional-data). The published antimalarial dose–response matrix data are available at the Tripod repository (https://tripod.nih.gov/matrix-client/rest/matrix/blocks/506/table)10. The published antiviral dose–response matrix data for Ebola treatment are available from the NCATS Matrix portal (https://matrix.ncats.nih.gov/matrix-client/rest/matrix/blocks/6324/table)35.
Code availability
The source code of the DECREASE prediction algorithm is freely available either at the tool website (http://decrease.fimm.fi/source_code) or GitHub (https://github.com/IanevskiAleksandr/DECREASE) to allow replication of the results and to compare or combine the cNMF algorithm with other prediction models. The source codes of the other algorithms are publicly available (web links and package versions are listed in Supplementary Table 3). The recent Dose model33 implementation was provided by request from its authors, A. Zimmer and U. Alon.
References
Webster, R. M. Combination therapies in oncology. Nat. Rev. Drug Discov. 15, 81–82 (2016).
Lehár, J. et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666 (2009).
O’Neil, J. et al. An unbiased oncology compound screen to identify novel combination strategies. Mol. Cancer Ther. 15, 1155–1162 (2016).
Radic-Sarikas, B. et al. Combinatorial drug screening identifies Ewing sarcoma-specific sensitivities. Mol. Cancer Ther. 16, 88–101 (2017).
Pelaia, G., Vatrella, A. & Maselli, R. The potential of biologics for the treatment of asthma. Nat. Rev. Drug Discov. 11, 958–972 (2012).
Chakradhar, S. All in one: researchers create combination drugs for diabetes and obesity. Nat. Med. 22, 694–696 (2016).
Worthington, R. J. & Melander, C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 31, 177–184 (2013).
Chandrasekaran, S. et al. Chemogenomics and orthology-based design of antibiotic combination therapies. Mol. Syst. Biol. 12, 872 (2016).
Borisy, A. A. et al. Systematic discovery of multicomponent therapeutics. Proc. Natl Acad. Sci. USA 100, 7977–7982 (2003).
Mott, B. T. et al. High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci. Rep. 5, 13891 (2015).
Mathews Griner, L. A. et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc. Natl Acad. Sci. USA 111, 2349–2354 (2014).
Janzen, W. P. Screening technologies for small molecule discovery: the state of the art. Chem. Biol. 21, 1162–1170 (2014).
He, L. et al. Patient-customized drug combination prediction and testing for T-cell prolymphocytic leukemia patients. Cancer Res. 78, 2407–2418 (2018).
Holbeck, S. L. et al. The national cancer institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res. 13, 3564–3576 (2017).
Roy, A. et al. Open access high throughput drug discovery in the public domain: a Mount Everest in the making. Curr. Pharm. Biotechnol. 11, 764–778 (2010).
Kurtz, S. E. et al. Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies. Proc. Natl Acad. Sci. USA 114, 7554–7563 (2017).
Yu, D. et al. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci. Rep. 5, 16991 (2015).
Horn, T. et al. High-order drug combinations are required to effectively kill colorectal cancer cells. Cancer Res. 76, 6950–6963 (2016).
Chou, A. et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut 67, 2142–2155 (2018).
Chou, Ting-Chao Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681 (2006).
Sun, W. et al. Rapid antimicrobial susceptibility test for identification of new therapeutics and drug combinations against multidrug-resistant bacteria. Emerg. Microbes Infect. 5, e116 (2016).
Shehata, M. et al. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood 116, 2513–2521 (2010).
Tan, X. et al. Systematic identification of synergistic drug pairs targeting HIV. Nat. Biotechnol. 30, 1125–1130 (2012).
Di Veroli, G. Y. et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32, 2866–2868 (2016).
Ianevski, A., He, L., Aittokallio, T. & Tang, J. SynergyFinder: a web application for analyzing drug combination dose–response matrix data. Bioinformatics 33, 2413–2415 (2017).
Loewe, S. The problem of synergism and antagonism of combined drugs. Arzneimiettelforschung 3, 286–290 (1953).
Bliss, C. I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615 (1939).
Berenbaum, M. C. What is synergy? Pharmacol. Rev. 41, 93–141 (1989).
Yadav, B. et al. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 13, 504–505 (2015).
Chevereau, G. & Bollenbach, T. Systematic discovery of drug interaction mechanisms. Mol. Syst. Biol. 11, 807 (2015).
Chou, T. C. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 70, 440–446 (2010).
Al-Lazikani, B. et al. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 30, 679–692 (2012).
Zimmer, A. et al. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl Acad. Sci. USA 113, 10442–10447 (2016).
Eziefula, A. C. Artesunate-mefloquine: a malaria treatment for African children? Lancet Infect. Dis. 16, 1086–1087 (2016).
Dyall, J. et al. Identification of combinations of approved drugs with synergistic activity against Ebola virus in cell cultures. J. Infect. Dis. 218, S672–S678 (2018).
Weinstein, Z. B. et al. Prediction of synergistic drug combinations. Curr. Opin. Syst. Biol. 4, 24–28 (2015).
Szwajda, A. et al. Systematic mapping of kinase addiction combinations in breast cancer cells by integrating drug sensitivity and selectivity profiles. Chem. Biol. 22, 1144–1155 (2015).
Mullard, A. Microfluidics platform lowers barrier to drug combination screening. Nat. Rev. Drug Discov. 17, 691–692 (2018).
Gautam, P. et al. Identification of selective cytotoxic and synthetic lethal drug responses in triple negative breast cancer cells. Mol. Cancer 15, 34 (2016).
Gadagkar, S. R. & Call, G. B. Computational tools for fitting the Hill equation to dose–response curves. J. Pharmacol. Toxicol. Methods 71, 68–76 (2015).
Hill, A. V. The possible effects of the aggregation of the molecules of hemoglobin on its dissociation curves. J. Physiol. 40, 4–7 (1910).
Ritz, C., Baty, F., Streibig, J. C. & Gerhard, D. Dose–response analysis using R. PLoS One 10, e0146021 (2015).
Wang, Y. X. et al. Non-negative matrix factorization: a comprehensive review. IEEE Trans. Knowl. Data Eng. 25, 1336–1353 (2013).
Venter, J. H. On estimation of the mode. Ann. Math. Stat. 38, 1446–1455 (1967).
Tang, J., Wennerberg, K. & Aittokallio, T. What is synergy? The Saariselkä agreement revisited. Front. Pharmacol. 6, 181 (2015).
Bischl, B. et al. mlrMBO: a modular framework for model-based optimization of expensive black-box functions. Preprint at https://arxiv.org/abs/1703.03373 (2017).
Chen, T. & Guestrin, C. Xgboost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016).
Acknowledgements
We thank the authors of the original publications (refs. 3,10,11,35) for making their drug combination data matrices publicly available. We thank the authors of the Dose model publication (ref. 33) for providing their MATLAB implementation and guaranteeing its appropriate use in this work, D. Bulanova (FIMM) for her valuable comments and discussions and O. Hansson (FIMM) for technical support with the web server. This work was funded by the Academy of Finland (grants nos. 272577 and 277293 to K.W. and 292611, 279163, 295504, 310507, 313267 and 326238 to T.A.), the European Union’s Horizon 2020 Research and Innovation Programme (ERA PerMed JAKSTAT-TARGET), the Cancer Society of Finland (T.A. and K.W.), Sigrid Jusélius Foundation (K.W. and T.A.) and Novo Nordisk Foundation (NNF17CC0027852 to K.W.). The FIMM High Throughput Biomedine Unit is supported financially by the University of Helsinki and Biocenter Finland (S.P. and J.S.).
Author information
Authors and Affiliations
Contributions
T.A., K.W. and A.I. designed and conceived the study. A.I. developed cNMF and implemented the web platform. A.I. and A.K.G. selected and tested various machine learning methods. P.G. performed the drug combination screening experiments. A.I., A.K.G. and A.K. managed data integration from various sources. A.I. and A.K. prepared the figures and tables for the manuscript. A.I., A.K.G. and T.A. wrote the manuscript. A.K., P.G., S.P., J.S. and K.W. critically reviewed and edited the manuscript. All authors reviewed and approved the final version of the manuscript. A.K.G. and P.G. contributed equally.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures, tables and methods
Rights and permissions
About this article
Cite this article
Ianevski, A., Giri, A.K., Gautam, P. et al. Prediction of drug combination effects with a minimal set of experiments. Nat Mach Intell 1, 568–577 (2019). https://doi.org/10.1038/s42256-019-0122-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-019-0122-4
This article is cited by
-
Multi-omics and immunogenomics analysis revealed PFKFB3 as a targetable hallmark and mediates sunitinib resistance in papillary renal cell carcinoma: in silico study with laboratory verification
European Journal of Medical Research (2024)
-
Harnessing machine learning to find synergistic combinations for FDA-approved cancer drugs
Scientific Reports (2024)
-
Performance evaluation of drug synergy datasets using computational intelligence approaches
Multimedia Tools and Applications (2024)
-
Targeting metabolism by B-raf inhibitors and diclofenac restrains the viability of BRAF-mutated thyroid carcinomas with Hif-1α-mediated glycolytic phenotype
British Journal of Cancer (2023)
-
Standardized assays to monitor drug sensitivity in hematologic cancers
Cell Death Discovery (2023)