Abstract
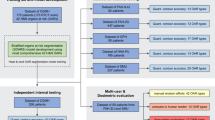
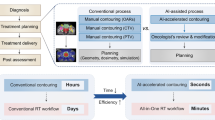
Radiation therapy is one of the most widely used therapies for cancer treatment. A critical step in radiation therapy planning is to accurately delineate all organs at risk (OARs) to minimize potential adverse effects to healthy surrounding organs. However, manually delineating OARs based on computed tomography images is time-consuming and error-prone. Here, we present a deep learning model to automatically delineate OARs in head and neck, trained on a dataset of 215 computed tomography scans with 28 OARs manually delineated by experienced radiation oncologists. On a hold-out dataset of 100 computed tomography scans, our model achieves an average Dice similarity coefficient of 78.34% across the 28 OARs, significantly outperforming human experts and the previous state-of-the-art method by 10.05% and 5.18%, respectively. Our model takes only a few seconds to delineate an entire scan, compared to over half an hour by human experts. These findings demonstrate the potential for deep learning to improve the quality and reduce the treatment planning time of radiation therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Because of patient privacy, access to the training data in Dataset 1 will be granted on a case by case basis on submission of a request to the corresponding authors. The availability of the train data is subject to review and approval by IRB. The test data in Dataset 1 is available for non-commercial research purpose at https://github.com/uci-cbcl/UaNet#Data. The CT images for Dataset 2 are freely available at https://doi.org/10.7937/K9/TCIA.2015.7AKGJUPZ and https://doi.org/10.7937/K9/TCIA.2017.8oje5q00; we provide the annotated dataset (freely available) for non-commercial use at https://github.com/uci-cbcl/UaNet#Data. Dataset 3 is freely available at http://www.imagenglab.com/newsite/pddca/.
Code availability
Code for the algorithm development, evaluation and statistical analysis is freely available for non-commercial research purposes (https://github.com/uci-cbcl/UaNet).
References
Lai, S.-Z. et al. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int. J. Radiat. Oncol. Biol. Phys. 80, 661–668 (2011).
Peng, G. et al. A prospective, randomized study comparing outcomes and toxicities of intensity-modulated radiotherapy vs. conventional two-dimensional radiotherapy for the treatment of nasopharyngeal carcinoma. Radiother. Oncol. 104, 286–293 (2012).
Bernier, J. et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N. Engl. J. Med. 350, 1945–1952 (2004).
Bonner, J. A. et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 11, 21–28 (2010).
Liu, W.-S. et al. Long-term results of intensity-modulated radiotherapy concomitant with chemotherapy for hypopharyngeal carcinoma aimed at laryngeal preservation. BMC Cancer 10, 102 (2010).
Prades, J.-M. et al. Randomized phase III trial comparing induction chemotherapy followed by radiotherapy to concomitant chemoradiotherapy for laryngeal preservation in T3M0 pyriform sinus carcinoma. Acta Otolaryngol. 130, 150–155 (2010).
Sun Yoon, M. et al. Concurrent chemoradiotherapy with cisplatin and fluorouracil for locally advanced hypopharyngeal carcinoma. Acta Otolaryngol. 128, 590–596 (2008).
Forastiere, A. A. et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N. Engl. J. Med. 349, 2091–2098 (2003).
Jones, A. S., Fish, B., Fenton, J. E. & Husband, D. J. The treatment of early laryngeal cancers (T1–T2 N0): surgery or irradiation? Head Neck J. Sci. Specialties Head Neck 26, 127–135 (2004).
Cooper, J. S. et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 350, 1937–1944 (2004).
Yeh, S.-A., Tang, Y., Lui, C.-C., Huang, Y.-J. & Huang, E.-Y. Treatment outcomes and late complications of 849 patients with nasopharyngeal carcinoma treated with radiotherapy alone. Int. J. Radiat. Oncol. Biol. Phys. 62, 672–679 (2005).
Hsiao, K.-Y. et al. Cognitive function before and after intensity-modulated radiation therapy in patients with nasopharyngeal carcinoma: a prospective study. Int. J. Radiat. Oncol. Biol. Phys. 77, 722–726 (2010).
Esteller, E., León, X., de Juan, M. & Quer, M. Delayed carotid blow-out syndrome: a new complication of chemoradiotherapy treatment in pharyngolaryngeal carcinoma. J. Laryngol. Otol. 126, 1189–1191 (2012).
Dietz, A. et al. Induction chemotherapy with paclitaxel and cisplatin followed by radiotherapy for larynx organ preservation in advanced laryngeal and hypopharyngeal cancer offers moderate late toxicity outcome (deLOS-I-trial). Eur. Arch. Otorhinolaryngol. 266, 1291–1300 (2009).
Fesneau, M. et al. Concomitant chemoradiotherapy using carboplatin, tegafur-uracil and leucovorin for stage III and IV head-and-neck cancer: results of gortec phase II study. Int. J. Radiat. Oncol. Biol. Phys. 76, 154–163 (2010).
Lee, D. S. et al. Long-term outcome and toxicity of hypofractionated stereotactic body radiotherapy as a boost treatment for head and neck cancer: the importance of boost volume assessment. Radiat. Oncol. 7, 85 (2012).
Machtay, M. et al. Factors associated with severe late toxicity after concurrent chemoradiation for locally advanced head and neck cancer: an RTOG analysis. J. Clin. Oncol. 26, 3582–3589 (2008).
Nguyen-Tan, P. F. et al. Randomized phase III trial to test accelerated versus standard fractionation in combination with concurrent cisplatin for head and neck carcinomas in the radiation therapy oncology group 0129 trial: long-term report of efficacy and toxicity. J. Clin. Oncol. 32, 3858–3866 (2014).
Daisne, J.-F. & Blumhofer, A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: a clinical validation. Radiat. Oncol. 8, 154 (2013).
Fortunati, V. et al. Tissue segmentation of head and neck CT images for treatment planning: a multiatlas approach combined with intensity modeling. Med. Phys. 40, 071905 (2013).
Duc, H. et al. Validation of clinical acceptability of an atlas-based segmentation algorithm for the delineation of organs at risk in head and neck cancer. Med. Phys. 42, 5027–5034 (2015).
Hoogeman, M. et al. Atlas-based auto-segmentation of CT images in head and neck cancer: what is the best approach? Int. J. Radiat. Oncol. Biol. Phys. 72, S591 (2008).
Levendag, P. et al. Atlas based auto-segmentation of CT images: clinical evaluation of using auto-contouring in high-dose, high-precision radiotherapy of cancer in the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 72, S401 (2008).
Qazi, A. A. et al. Auto-segmentation of normal and target structures in head and neck CT images: a feature-driven model-based approach. Med. Phys. 38, 6160–6170 (2011).
Sims, R. et al. A pre-clinical assessment of an atlas-based automatic segmentation tool for the head and neck. Radioth. Oncol. 93, 474–478 (2009).
Teguh, D. N. et al. Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 81, 950–957 (2011).
Thomson, D. et al. Evaluation of an automatic segmentation algorithm for definition of head and neck organs at risk. Radiat. Oncol. 9, 173 (2014).
Walker, G. V. et al. Prospective randomized double-blind study of atlas-based organ-at-risk autosegmentation-assisted radiation planning in head and neck cancer. Radioth. Oncol. 112, 321–325 (2014).
Voet, P. W. et al. Does atlas-based autosegmentation of neck levels require subsequent manual contour editing to avoid risk of severe target underdosage? a dosimetric analysis. Radioth. Oncol. 98, 373–377 (2011).
Isambert, A. et al. Evaluation of an atlas-based automatic segmentation software for the delineation of brain organs at risk in a radiation therapy clinical context. Radioth. Oncol. 87, 93–99 (2008).
Fritscher, K. D. et al. Automatic segmentation of head and neck CT images for radiotherapy treatment planning using multiple atlases, statistical appearance models, and geodesic active contours. Med. Phys. 41, 051910 (2014).
Commowick, O., Grégoire, V. & Malandain, G. Atlas-based delineation of lymph node levels in head and neck computed tomography images. Radioth. Oncol. 87, 281–289 (2008).
Verhaart, R. F. et al. The relevance of MRI for patient modeling in head and neck hyperthermia treatment planning: a comparison of CT and CT–MRI based tissue segmentation on simulated temperature. Med. Phys. 41, 123302 (2014).
Wachinger, C., Fritscher, K., Sharp, G. & Golland, P. Contour-driven atlas-based segmentation. IEEE Trans. Med. Imaging 34, 2492–2505 (2015).
Fortunati, V. et al. Automatic tissue segmentation of head and neck MR images for hyperthermia treatment planning. Phys. Med. Biol. 60, 6547–6562 (2015).
Zhang, T., Chi, Y., Meldolesi, E. & Yan, D. Automatic delineation of on-line head-and-neck computed tomography images: toward on-line adaptive radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 68, 522–530 (2007).
Kosmin, M. et al. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radioth. Oncol. 135, 130–140 (2019).
Ren, X. et al. Interleaved 3D-CNNs for joint segmentation of small-volume structures in head and neck CT images. Med. Phys. 45, 2063–2075 (2018).
Močnik, D. et al. Segmentation of parotid glands from registered CT and MR images. Phys. Med 52, 33–41 (2018).
Fritscher, K. et al. Deep neural networks for fast segmentation of 3D medical images. In International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Ourselin, S., Joskowicz, L., Sabuncu, M. R., Unal, G. & Wells, W.) 158–165 (Springer, 2016).
Ibragimov, B. & Xing, L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med. Phys. 44, 547–557 (2017).
Tam, C. et al. Automated delineation of organs-at-risk in head and neck CT images using multi-output support vector regression. In Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging Vol. 10578 (eds Gimi, B. & Krol, A.) 1057824 (International Society for Optics and Photonics, 2018).
Wu, X. et al. Auto-contouring via automatic anatomy recognition of organs at risk in head and neck cancer on CT images. In Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling Vol. 10576 (eds Gimi, B. & Krol, A.) 1057617 (International Society for Optics and Photonics, 2018).
Tong, Y. et al. Hierarchical model-based object localization for auto-contouring in head and neck radiation therapy planning. In Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging Vol. 10578 (eds Gimi, B. & Krol, A.) 1057822 (International Society for Optics and Photonics, 2018).
Pednekar, G. V. et al. Image quality and segmentation. In Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling Vol. 10576 (eds Gimi, B. & Krol, A.) 105762N (International Society for Optics and Photonics, 2018).
Wang, Z. et al. Hierarchical vertex regression-based segmentation of head and neck CT images for radiotherapy planning. IEEE Trans. Image Process. 27, 923–937 (2018).
Cardenas, C. E. et al. Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int. J. Radiat. Oncol. Biol. Phys. 101, 468–478 (2018).
Nikolov, S. et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. Preprint at https://arxiv.org/abs/1809.04430 (2018).
Hänsch, A. et al. Comparison of different deep learning approaches for parotid gland segmentation from CT images. In Medical Imaging 2018: Computer-Aided Diagnosis Vol. 10575 (eds Gimi, B. & Krol, A.) 1057519 (International Society for Optics and Photonics, 2018).
Tong, N., Gou, S., Yang, S., Ruan, D. & Sheng, K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med. Phys. 45, 4558–4567 (2018).
Zhu, W. et al. Anatomynet: Deep 3D squeeze-and-excitation U-Nets for fast and fully automated whole-volume anatomical segmentation. Preprint at https://arxiv.org/abs/1808.05238 (2018).
Men, K. et al. More accurate and efficient segmentation of organs-at-risk in radiotherapy with convolutional neural networks cascades. Med. Phys. 46, 286–292 (2019).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer, 2015).
Milletari, F., Navab, N. & Ahmadi, S.-A. V-net: fully convolutional neural networks for volumetric medical image segmentation. In 2016 Fourth International Conference on 3D Vision (3DV) 565–571 (IEEE, 2016).
Ren, S., He, K., Girshick, R. & Sun, J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39, 1137–1149 (2016).
Clark, K. et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013).
Brouwer, C. L. et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother. Oncol. 117, 83–90 (2015).
Bosch, W. R., Straube, W. L., Matthews, J. W. & Purdy, J. A. Head–Neck Cetuximab (Cancer Imaging Archive, 2015); https://wiki.cancerimagingarchive.net/x/xwxp
Vallières, M. et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 7, 10117 (2017).
Raudaschl, P. F. et al. Evaluation of segmentation methods on head and neck CT: auto-segmentation challenge 2015. Med. Phys. 44, 2020–2036 (2017).
Huttenlocher, D. P., Rucklidge, W. J. & Klanderman, G. A. Comparing images using the Hausdorff distance under translation. In Proceedings of 1992 IEEE Computer Society Conference on Computer Vision and Pattern Recognition 654–656 (IEEE, 1992).
van der Veen, J. et al. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother. Oncol. 138, 68–74 (2019).
Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision 1440–1448 (IEEE, 2015).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE, 2018).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. In 2017 IEEE International Conference on Computer Vision (ICCV) 2980–2988 (IEEE, 2017).
Lin, T.-Y. et al. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2117–2125 (IEEE, 2017).
Acknowledgements
We acknowledge helpful discussions with D. Chow, W. Zhu and S. Vang. This work was supported by grants from the National Natural Science Foundation of China (no. 81872547), the Natural Science Foundation of Shanghai (no. 18ZR1430800), the Shanghai Jiao Tong University Medical and Engineering Collaborative Research Fund Project (no. YG2016MS24) and Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (no. 20181713).
Author information
Authors and Affiliations
Contributions
Y.L. and X.X. initiated the project. H.T. and X.X. designed the model architecture. H.T., Yang Liu and X.C. conducted validation experiments. X.C., S.Y., G.Z., Y.X., T.C. and Yong Liu created the dataset. H.T., X.C., Yong Liu and X.X. wrote the paper. Z.L., J.Y. and M.Y. contributed to software development.
Corresponding authors
Ethics declarations
Competing interests
H.T. interned with and received a stipend from DeepVoxel. X.X. is an advisor to DeepVoxel. Y.L., Z.L., J.Y. and M.Y. are employees of DeepVoxel. None of the other authors declare competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures, Supplementary tables, Supplementary methods and Supplementary discussion.
Rights and permissions
About this article
Cite this article
Tang, H., Chen, X., Liu, Y. et al. Clinically applicable deep learning framework for organs at risk delineation in CT images. Nat Mach Intell 1, 480–491 (2019). https://doi.org/10.1038/s42256-019-0099-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-019-0099-z
This article is cited by
-
Mandible segmentation from CT data for virtual surgical planning using an augmented two-stepped convolutional neural network
International Journal of Computer Assisted Radiology and Surgery (2023)
-
Comprehensive and clinically accurate head and neck cancer organs-at-risk delineation on a multi-institutional study
Nature Communications (2022)
-
Annotation-efficient deep learning for automatic medical image segmentation
Nature Communications (2021)
-
Automatic mandible segmentation from CT image using 3D fully convolutional neural network based on DenseASPP and attention gates
International Journal of Computer Assisted Radiology and Surgery (2021)