Abstract
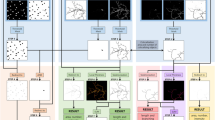
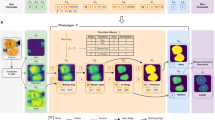
The segmentation of images is a common task in a broad range of research fields. To tackle increasingly complex images, artificial intelligence-based approaches have emerged to overcome the shortcomings of traditional feature detection methods. Owing to the fact that most artificial intelligence research is made publicly accessible and programming the required algorithms is now possible in many popular languages, the use of such approaches is becoming widespread. However, these methods often require data labelled by the researcher to provide a training target for the algorithms to converge to the desired result. This labelling is a limiting factor in many cases and can become prohibitively time consuming. Inspired by the ability of cycle-consistent generative adversarial networks to perform style transfer, we outline a method whereby a computer-generated set of images is used to segment the true images. We benchmark our unsupervised approach against a state-of-the-art supervised cell-counting network on the VGG Cells dataset and show that it is not only competitive but also able to precisely locate individual cells. We demonstrate the power of this method by segmenting bright-field images of cell cultures, images from a live/dead assay of C. elegans, and X-ray computed tomography of metallic nanowire meshes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Our tensorflow implementation of the cycleGAN with our novel histogram discriminator, all of the synthetic-data-generating scripts and the analysis and post-processing scripts are available at https://github.com/UDCTGAN/UDCT.
Data availability
The bright-field dataset of primary cortical neurons with ground-truth labels, the annotated C. elegans dataset, the dataset of X-ray computed tomography of silver nanowires, all trained networks and the scripts for creating the synthetic datasets are available at https://downloads.lbb.ethz.ch.
References
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Badrinarayanan, V., Kendall, A. & Cipolla, R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017).
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D u-net: learning dense volumetric segmentation from sparse annotation. In Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Ourselin, S., Joskowicz, L., Sabuncu, M. R., Wells, W. & Unal, G.) 424–432 (Springer, 2016).
Falk, T. et al. U-net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2018).
Jégou, S., Drozdzal, M., Vazquez, D., Romero, A. & Bengio, Y. The one hundred layers tiramisu: Fully convolutional densenets for semantic segmentation. In Proc. Conference on Computer Vision and Pattern Recognition Workshops 1175–1183 (IEEE, 2017).
Kamal, U. et al. Lung cancer tumor region segmentation using recurrent 3D-DenseUNet. Preprint at https://arXiv.org/abs/1812.01951 (2018).
Konopczyński, T., Kröger, T., Zheng, L. & Hesser, J. Instance segmentation of fibers from low resolution CT scans via 3D deep embedding learning. Preprint at https://arXiv.org/abs/1511.00561 (2019).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proc. IEEE International Conference on Computer Vision 2223–2232 (IEEE, 2017).
Goodfellow, I. et al. Generative adversarial nets. In Proc. Advances in Neural Information Processing Systems 2672–2680 (NIPS, 2014).
Karras, T., Laine, S. & Aila, T. A style-based generator architecture for generative adversarial networks. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 4401–4410 (IEEE, 2019).
Haering, M., Grosshans, J., Wolf, F. & Eule, S. Automated segmentation of epithelial tissue using cycle-consistent generative adversarial networks. Preprint at https://doi.org/10.1101/311373 (2018).
Zhang, Z., Yang, L. & Zheng, Y. Translating and segmenting multimodal medical volumes with cycle-and shapeconsistency generative adversarial network. In Proc. Conference on Computer Vision and Pattern Recognition 9242–9251 (IEEE, 2018).
Xu, Z., Moro, C. F., Bozóky, B. & Zhang, Q. GAN-based virtual re-staining: a promising solution for whole slide image analysis. Preprint at https://arXiv.org/abs/1901.04059 (2019).
Huo, Y. et al. Adversarial synthesis learning enables segmentation without target modality ground truth. In Proc. 15th International Symposium on Biomedical Imaging 1217–1220 (IEEE, 2018).
Chen, C., Dou, Q., Chen, H. & Heng, P.-A. Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest X-ray segmentation. In International Workshop on Machine Learning in Medical Imaging 143–151 (Springer, 2018).
Fu, C. et al. Three dimensional fluorescence microscopy image synthesis and segmentation. In Proc. IEEE Conference on Computer Vision and Pattern Recognition Workshops 2221–2229 (2018).
Mahmood, F. et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. In IEEE Transactions on Medical Imaging (IEEE, 2019).
Lempitsky, V. & Zisserman, A. VGG Cell dataset from learning to count objects in images. In Advances in Neural Information Processing Systems (NIPS, 2010).
Cohen, J. P., Boucher, G., Glastonbury, C. A., Lo, H. Z. & Bengio, Y. Count-ception: counting by fully convolutional redundant counting. In Proc. International Conference on Computer Vision 18–26 (IEEE, 2017).
Moy, T. I. et al. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 4, 527–533 (2009).
Ljosa, V., Sokolnicki, K. L. & Carpenter, A. E. Annotated high-throughput microscopy image sets for validation. Nat. Methods 9, 637 (2012).
Wählby, C. et al. An image analysis toolbox for high-throughput C. elegans assays. Nat. Methods 9, 714 (2012).
Stampanoni, M. et al. in Developments in X-ray Tomography V, Vol. 6318, 63180M (International Society for Optics and Photonics, 2006).
Stampanoni, M. et al. Phase-contrast tomography at the nanoscale using hard x rays. Phys. Rev. B 81, 140105 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676 (2012).
Carpenter, A. E. et al. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Ulyanov, D., Vedaldi, A. & Lempitsky, V. Instance normalization: the missing ingredient for fast stylization. Preprint at http://arxiv.org/abs/1607.08022v3 (2017).
Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 1125–1134 (IEEE, 2017).
Salimans, T. et al. Improved techniques for training GANs. In Proc. Advances in Neural Information Processing Systems 2234–2242 (NIPS, 016).
Risser, E., Wilmot, P. & Barnes, C. Stable and controllable neural texture synthesis and style transfer using histogram losses. Preprint at http://arxiv.org/abs/1701.08893v2 (2017).
Yosinski, J., Clune, J., Bengio, Y. & Lipson, H. How transferable are features in deep neural networks? In Proc. Advances in Neural Information Processing Systems 3320–3328 (NIPS, 2014).
Acknowledgements
The research was financed by ETH Zurich, the Swiss National Science Foundation (project no. 165651), the Swiss Data Science Center and a FreeNovation grant. We also acknowledge the Paul Scherrer Institute, Villigen, Switzerland, for provision of synchrotron radiation beamtime at beamline TOMCAT of the Swiss Light Source and D. Krüger, E. Konukoglu, S. Stoma, G. Csúcs, A. J. Rzeplela and S. F. Nrrelykke for the insightful and valuable feedback on earlier versions of the manuscript.
Author information
Authors and Affiliations
Contributions
S.J.I. and C.F. designed the research project. S.J.I. and C.F. implemented and tested the code. S.G. performed the neuronal dataset acquisition. A.M.R., H.H., F.S., A.B., M.S. and C.F. carried out the nanowire dataset acquisition. C.F. trained the network on the nanowire and cell VGG dataset. S.J.I. and C.F. trained the network on the C. elegans and dead versus live neuronal dataset. S.J.I. trained the network on the colour-coding of the live dataset. S.J.I., K.P. and C.F. performed the pre- and post-processing of the data. S.J.I., A.M.R., C.F. and S.G. labelled the neuronal datasets. A.M.R., S.G., H.H., A.B., M.S., K.P. and J.V. gave critical input and discussion. All co-authors reviewed the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Description of videos, network architecture, datasets and processing steps.
Supplementary Video 1
Effect of background noise on synthetic image generation.
Supplementary Video 2
Effect of object density on synthetic image generation.
Supplementary Video 3
Effect of object shape on synthetic image generation.
Supplementary Video 4
Effect of object size on synthetic image generation.
Rights and permissions
About this article
Cite this article
Ihle, S.J., Reichmuth, A.M., Girardin, S. et al. Unsupervised data to content transformation with histogram-matching cycle-consistent generative adversarial networks. Nat Mach Intell 1, 461–470 (2019). https://doi.org/10.1038/s42256-019-0096-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-019-0096-2
This article is cited by
-
Multi-focus light-field microscopy for high-speed large-volume imaging
PhotoniX (2022)
-
Deep learning-based segmentation of lithium-ion battery microstructures enhanced by artificially generated electrodes
Nature Communications (2021)
-
Predicting carbon nanotube forest attributes and mechanical properties using simulated images and deep learning
npj Computational Materials (2021)
-
Unsupervised content-preserving transformation for optical microscopy
Light: Science & Applications (2021)
-
Scaffold-A549: A Benchmark 3D Fluorescence Image Dataset for Unsupervised Nuclei Segmentation
Cognitive Computation (2021)