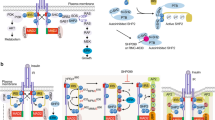
Abstract
Insulin signaling is essential for glucose metabolism, and insulin decreases insulin receptor (InsR) levels in a dose-dependent and time-dependent manner. However, the regulatory mechanisms of InsR reduction upon insulin stimulation remain poorly understood. Here, we show that Eph receptor B4 (EphB4), a tyrosine kinase receptor that modulates cell adhesion and migration, can bind directly to InsR, and this interaction is markedly enhanced by insulin. Due to the adaptor protein 2 (Ap2) complex binding motif in EphB4, the interaction of EphB4 and InsR facilitates clathrin-mediated InsR endocytosis and degradation in lysosomes. Hepatic overexpression of EphB4 decreases InsR and increases hepatic and systemic insulin resistance in chow-fed mice, whereas genetic or pharmacological inhibition of EphB4 improve insulin resistance and glucose intolerance in obese mice. These observations elucidate a role for EphB4 in insulin signaling, suggesting that EphB4 might represent a therapeutic target for the treatment of insulin resistance and type 2 diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Primer sequences for qPCR are listed in Supplementary Tables 1 and 2. The sequence information for designing EPHB4 shRNAs is listed in Supplementary Table 3. The clinical features of the patients are listed in Supplementary Table 4. All source data for immunoblotting are shown in Supplementary Fig. 1. Other source data in this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Olefsky, J. M. & Glass, C. K. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246 (2010).
Saltiel, A. R. & Olefsky, J. M. Thiazolidinediones in the treatment of insulin resistance and type II diabetes. Diabetes 45, 1661–1669 (1996).
Li, P. et al. LTB4 promotes insulin resistance in obese mice by acting on macrophages, hepatocytes and myocytes. Nat. Med. 21, 239–247 (2015).
Shanik, M. H. et al. Insulin resistance and hyperinsulinemia: is hyperinsulinemia the cart or the horse? Diabetes Care 31, S262–S268 (2008).
Corkey, B. E. Banting lecture 2011: hyperinsulinemia: cause or consequence? Diabetes 61, 4–13 (2012).
Marbán, S. L. & Roth, J. in Lessons from Animal Diabetes VI Vol. 6 (ed Shafrir, E.) (Birkhäuser Boston, 1996).
Destefano, M. B., Stern, J. S. & Castonguay, T. W. Effect of chronic insulin administration on food intake and body weight in rats. Physiol. Behav. 50, 801–806 (1991).
Martin, C., Desai, K. S. & Steiner, G. Receptor and postreceptor insulin resistance induced by in vivo hyperinsulinemia. Can. J. Physiol. Pharmacol. 61, 802–807 (1983).
Rizza, R. A., Mandarino, L. J., Genest, J., Baker, B. A. & Gerich, J. E. Production of insulin resistance by hyperinsulinaemia in man. Diabetologia 28, 70–75 (1985).
Gavin, J. R.III, Roth, J., Neville, D. M.Jr., De Meyts, P. & Buell, D. N. Insulin-dependent regulation of insulin receptor concentrations: a direct demonstration in cell culture. Proc. Natl Acad. Sci. USA 71, 84–88 (1974).
De Meyts, P., Roth, J., Neville, D. M.Jr., Gavin, J. R.III & Lesniak, M. A. Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem. Biophys. Res. Commun. 55, 154–161 (1973).
Kobayashi, M. & Olefsky, J. M. Effect of experimental hyperinsulinemia on insulin binding and glucose transport in isolated rat adipocytes. Am. J. Physiol. 235, E53–E62 (1978).
Goldfine, I. D. et al. Decreased binding of insulin to its receptors in rats with hormone induced insulin resistance. Biochem. Biophys. Res. Commun. 53, 852–857 (1973).
Czech, M. P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 23, 804–814 (2017).
Després, J.-P. & Lemieux, I. Abdominal obesity and metabolic syndrome. Nature 444, 881–887 (2006).
Hubbard, S. R. & Till, J. H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 69, 373–398 (2000).
Clague, M. J. & Urbé, S. Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 (2010).
Choi, E., Zhang, X., Xing, C. & Yu, H. Mitotic checkpoint regulators control insulin signaling and metabolic homeostasis. Cell 166, 567–581 (2016).
Choi, E. et al. Mitotic regulators and the SHP2-MAPK pathway promote IR endocytosis and feedback regulation of insulin signaling. Nat. Commun. 10, 1473 (2019).
Ansarullah et al. Inceptor counteracts insulin signalling in β-cells to control glycaemia. Nature 590, 326–331 (2021).
Hotamisligil, G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006).
Giorgio, C. et al. Lithocholic acid is an Eph-ephrin ligand interfering with Eph-kinase activation. PLoS ONE 6, e18128 (2011).
Martiny-Baron, G. et al. The small molecule specific EphB4 kinase inhibitor NVP-BHG712 inhibits VEGF driven angiogenesis. Angiogenesis 13, 259–267 (2010).
Li, P. et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell 147, 815–826 (2011).
Li, P. et al. Hematopoietic-derived galectin-3 causes cellular and systemic insulin resistance. Cell 167, 973–984.e12 (2016).
Reznik, Y. & Cohen, O. Insulin pump for type 2 diabetes: use and misuse of continuous subcutaneous insulin infusion in type 2 diabetes. Diabetes Care 36, S219–S225 (2013).
Virtue, S. & Vidal-Puig, A. GTTs and ITTs in mice: simple tests, complex answers. Nat. Metab. 3, 883–886 (2021).
Li, P. et al. Functional heterogeneity of CD11c-positive adipose tissue macrophages in diet-induced obese mice. J. Biol. Chem. 285, 15333–15345 (2010).
He, W. et al. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl Acad. Sci. USA 100, 15712–15717 (2003).
Acknowledgements
We thank Jerrold M. Olefsky at the University of California San Diego for providing advice. We thank Xiaowei Zhang at Peking University for providing the albumin-cre mice. This work was supported by grants from the National Key R&D Program of China (2017YFA0205400), Beijing Outstanding Young Scientist Program (BJJWZYJH01201910023028), the National Natural Science Foundation China (81700767, 81622010, 81770800, 81874316 and 81703588), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2021-I2M-1-016, 2016-I2M-1-011, 2016-I2M-3-008 and 2017-I2M-1-008), the CAMS Central Public-interest Scientific Institution Basal Research Fund (2017RC31009 and 2018PT35004) and the Drug Innovation Major Project from the National Science and Technology Department (2018ZX09711001-003-005 and 2018ZX09711001-003-001).
Author information
Authors and Affiliations
Contributions
X.L. performed the overall experiments and analyzed the data. K.W. and J.C. isolated the primary hepatocytes from mice and performed the gluconeogenesis assay. S.H. and Q.J. performed the clamp experiment. L.K., Q.J., Z.W., C.M., Q.Z. and Y.H. assisted with the tissue collection and qPCR experiment. T.Y., Z.S., H.Z. and Y.L. assisted with the animal and cell experiments. P.L. conceived the project and directed the research. Z. Huang, Z. Hu and B.C. provided constructive advice. X.L., B.C. and P.L. wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare.
Peer review
Peer review information
Nature Metabolism thanks Heiko Lickert and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HFD induced hyperinsulinemia and InsR protein reduction.
a-g, Body weight curve (a, n = 3 mice), blood insulin curve (b, n = 3 mice), plasma glucose curve (c, n = 3 mice), and hepatic InsR protein levels (d, e, n = 3 mice) in C57BL/6J mice during HFD feeding. f, InsR protein level in epi-WAT of HFD-fed mice (age: 24 weeks, HFD: 16 weeks, n = 3 mice). g, InsR protein level in muscles of HFD-fed mice (n = 3 mice). h, Hepatic InsR mRNA levels (n = 3 mice). For statistical analysis in a-c and e-h, comparisons were versus related controls, using two-sided Student’s t-test. p values in a denoted by *** were p < 0.0001. Values are expressed as the mean ± SEM. NC, normal chow; HFD, high fat diet.
Extended Data Fig. 2 Insulin decreased InsR protein levels.
a-c, Dose-dependent effect of insulin (6 h) on InsR protein levels in primary hepatocytes from C57BL/6J mice. n = 3 cells examined over 2 independent experiments in b, c. d-f, Time-dependent effect of insulin (100 nM) on InsR protein levels in primary hepatocytes from C57BL/6J mice. n = 3 cells examined over 2 independent experiments in e, f. For statistical analysis in b-c and e-f, comparisons were versus related controls, using two-sided Student’s t-test. p values in b denoted by asterisks (from left to right): p = 0.020, p = 0.024, p = 0.012, p = 0.0043. p values in c denoted by asterisks (from left to right): p = 0.033, p = 0.021, p = 0.012, p = 0.013, p = 0.0014. p values in e denoted by asterisks (from left to right): p = 0.010, p = 0.0053, p = 0.0020. p values in f denoted by asterisks (from left to right): p = 0.013, p = 0.031, p = 0.014, p = 0.029, p = 0.043. Values are presented as the mean ± SEM.
Extended Data Fig. 3 EphB4 interacted with InsR.
a, The identified peptide from EPHB4 by mass spectrum in INSR interaction protein screening in HepG2 cells. The b- and y-type product ions are marked on the mass spectrum and also illustrated in the peptide sequence shown above. b, c, Expression pattern of EphA and EphB families in C57BL/6 J mice by semiquantitative PCR. d, Interactions of homologous proteins of EPHB4 with InsR in HepG2 cells. e, Interaction of EPHB4 with other tyrosine kinases in HepG2 cells. f, Interaction of EPHB4 with IGF1R in HepG2 cells. g, h, Interaction of EPHB4-HA and Flag-INSR in HEK293T cells after Flag-INSR or EPHB4-HA was immunoprecipitated. i, Co-localization of EPHB4 and INSR detected using endogenous antibodies. Pearson’s R value: 0.57. j, Direct binding of EPHB4 and InsR in a pull-down assay using purified proteins. k, Diagrammatic sketch of the InsR protein structure. The short line indicates the truncated mutants of the full-length InsR protein. l, Interaction of EPHB4 with INSR domains with INSR truncation mutants. m, Diagrammatic sketch of the EPHB4 protein structure. The short line indicates the mutants in which part of the sequence was deleted from the full-length EPHB4 protein. n, Interaction of INSR with EPHB4 domains in EPHB4 deletion mutants. o, p, Interaction of INSR with EPHB4 (WT) and its kinase-dead mutant (KD) in HepG2 cells. n = 3 independent experiments in p. For statistical analysis in p, comparisons were versus related controls, using two-sided Student’s t-test. Values are presented as the mean ± SEM.
Extended Data Fig. 4 EphB4 expression pattern.
a, b, EphB4 mRNA (a, n = 3 mice) and protein (b, n = 3 mice) levels in the livers of HFD-fed mice. EPHB4 proteins were detected in the same samples used to detect Actin levels in Extended Data Fig. 1d. c, d, EPHB4 mRNA level in HepG2 cells (c, n = 5 biologically independent cells) and primary hepatocytes (d, n = 4 biologically independent cells) stimulated with insulin (100 nM). For statistical analysis in a-d, comparisons were versus related controls, using two-sided Student’s t-test. Values are presented as the mean ± SEM. NC, normal chow; HFD, high fat diet.
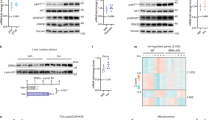
Extended Data Fig. 5 EphB4 promoted InsR degradation.
a, b, InsR protein levels upon insulin treatment and EPHB4 overexpression in primary hepatocytes. n = 3 biologically independent cells in b. c, d, Plasma membrane INSR protein levels were detected in control and EPHB4-overexpressing HepG2 cells by measuring surface biotinylation. n = 3 independent experiments in d. e, Body weight of Ad-EPHB4 mice. Age: 8 weeks. n = 11 mice. f, Plasma AST levels in Ad-EPHB4 mice. n = 11 mice. g, Plasma ALT levels in Ad-EPHB4 mice. n = 11 mice. h, Plasma insulin levels in in Ad-EPHB4 mice. n = 11 mice. i, InsR mRNA level in the Ad-EPHB4 mouse liver. n = 11 mice. j, k, Levels of ER stress-related proteins in EPHB4-overexpressing primary hepatocytes (i, n = 3 biologically independent cells) and livers (j, n = 6 mice). l, Protein synthesis assay using O-propargyl-puromycin (OPP) labeling. For statistical analysis in b and d-i, comparisons were versus related controls, using two-sided Student’s t-test. Values are presented as the mean ± SEM.
Extended Data Fig. 6 EphB4 promoted InsR degradation in the lysosome.
a, Effect of EPHB4 on INSR ubiquitination in HepG2 cells. b, c, Effect of proteasome inhibitors on EPHB4-induced INSR degradation. n = 3 biologically independent cell samples in c. d, Colocalization of EPHB4, INSR and Clathrin light chain (Clta) in HepG2 cells. Enlarged images are indicated by the white frames. Pearson’s R value for the colocalization of INSR with Clta: 0.65. e, Effect of EPHB4 overexpression on the interaction between InsR and the early endosome marker EEA1. f, Effect of EPHB4 overexpression on the interaction between INSR and the late endosome marker RAB7. g, Effect of EPHB4 overexpression on the interaction between INSR and the recycling endosome marker RAB11. h, Colocalization of EPHB4, INSR and RAB11 in HepG2 cells. Enlarged images were indicated by the white frames. Pearson’s R value for the colocalization of INSR with RAB11: 0.38. i, Interaction of AP2M1 with EPHB4 domains in EPHB4 truncation mutants. j, Ap2 binding motif sequence. k, Predicted Ap2 binding motifs in EPHB4. l, Statistical analysis of the rescue effect of mutants in the Ap2 binding motifs in EPHB4 on INSR degradation in HepG2 cells. n = 4 biologically independent cell samples. For statistical analysis in c and l, comparisons were versus related controls, using two-sided Student’s t-test. Values are expressed as the mean ± SEM.
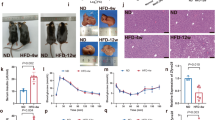
Extended Data Fig. 7 Lipids and inflammatory profiles in EphB4 LKO mice.
a, b, EPHB4 knockdown by two specific shRNAs in HepG2 cells. n = 4 biologically independent cell samples. c, EphB4 mRNA levels determined using qPCR in tissues from EphB4 LKO mice fed NC. Age: 5 weeks, n = 7 mice. d, e, Hepatic EphB4 and InsR protein levels in EphB4 LKO mice. n = 8 mice. f, Body weight of EphB4 LKO mice fed NC. n = 10 mice. g, GTT of EphB4 LKO mice fed NC. n = 10 mice. h, ITT of EphB4 LKO mice fed NC. n = 10 mice. i, Body weight of EphB4 LKO mice fed an HFD. n = 10 mice. j, Expression of inflammation-related genes in the liver of EphB4 LKO mice fed an HFD. n = 8 mice. k, Expression of inflammation-related genes in Epi-WAT of EphB4 LKO mice fed an HFD. n = 8 mice. For statistical analysis in b-c and e-k, comparisons were versus related controls, using two-sided Student’s t-test. Values are presented as the mean ± SEM. NC, normal chow.
Extended Data Fig. 8 Inflammatory profiles in db/db-EphB4 LKO mice.
a, Body weight of db/db-EphB4 LKO mice (n = 12 mice). b, Hepatic EphB4 protein levels in db/db-EphB4 LKO mice (n = 3 mice). c, Liver weight of db/db-EphB4 LKO mice (n = 9 mice). d, Liver index of db/db-EphB4 LKO mice (n = 9 mice). e, Epi-WAT mass of db/db-EphB4 LKO mice (n = 9 mice). f, Epi-WAT index of db/db-EphB4 LKO mice (n = 9 mice). g, Inflammatory gene expression in the livers of db/db-EphB4 LKO mice (n = 9 mice). h to j, Time-dependent effect of insulin (100 nM) on InsR protein levels in primary hepatocytes from EphB4 fl/fl and EphB4 LKO mice. n = 4 cells examined over 2 independent experiments. For statistical analysis in a-g and i-j, comparisons were versus related controls, using two-sided Student’s t-test. Values are expressed as the mean ± SEM.
Extended Data Fig. 9 A pharmacological EphB4 inhibitor improved metabolism in mice.
a, b, c, Effect of LCA on the insulin-stimulated increase in p-AKT levels in HepG2 cells (a, b, n = 3 biologically independent cell samples) and primary hepatocytes (c). d, e, Gluconeogenesis in primary hepatocytes from HFD-fed mice (d, n = 4 biologically independent cell samples) or db/db mice (e, n = 3 biologically independent cell samples) treated with LCA. f, Body weight of db/db mice grouped for drug administration (age: 8 weeks, n = 10 mice). g, Fasting plasma glucose level in db/db mice grouped for drug administration (n = 10 mice). h, Percent decrease in glucose levels after the insulin injection and drug administration (n = 10 mice). i, GTT of LCA-treated db/db mice (n = 9 mice). j, ITT of LCA-treated db/db mice (n = 9 mice). k, Serum TG levels in NVP-BHG712- or LCA-treated db/db mice (n = 9 mice). l-n, InsR protein level (n = 7 mice) and p-Akt levels (n = 3 mice) in the livers of LCA-treated db/db mice. For statistical analysis in b, d-e, i-k and m-n, comparisons were versus related controls, using two-sided Student’s t-test. For statistical analysis in f-h, comparisons were made for the three groups, using one-way ANOVA. p values in i denoted by asterisks (from left to right): p = 0.0069, p = 0.047, p = 0.0032, p = 0.0076, p = 0.0040. p values in j denoted by asterisks (from left to right): p = 0.022, p = 0.047, p = 0.042. Values are expressed as the mean ± SEM.
Extended Data Fig. 10 EphB4 shRNA improved metabolism in mice.
a, Body weight of db/db mice injected with the EphB4 shRNA (ctr shRNA n = 8 mice, EphB4 shRNA n = 14 mice). b, Hepatic EphB4 mRNA levels in EphB4 shRNA-injected mice (ctr shRNA n = 8 mice, EphB4 shRNA n = 14 mice). c, Hepatic EphB4 mRNA levels in EphB4 shRNA-injected mice. (ctr shRNA n = 6 mice, EphB4 shRNA n = 4 mice). d-e, p-Akt levels in the livers of EphB4 shRNA-injected mice after the insulin (0.75 U/kg) injection (age: 10 weeks, n = 3 mice in e). f, Gluconeogenesis assay in primary hepatocytes from EphB4 shRNA-injected mice (n = 5 biologically independent cell samples). g, Serum TG levels in EphB4 shRNA-injected mice (ctr shRNA n = 8 mice, EphB4 shRNA n = 14 mice). h, Graphic summary on insulin receptor degradation in the liver through EphB4. For statistical analysis in b-d and f-g, comparisons were versus related controls, using two-sided Student’s t-test. Values are expressed as the mean ± SEM.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Unprocessed western blots and gels.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Wang, K., Hou, S. et al. Insulin induces insulin receptor degradation in the liver through EphB4. Nat Metab 4, 1202–1213 (2022). https://doi.org/10.1038/s42255-022-00634-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-022-00634-5
This article is cited by
-
ZFYVE28 mediates insulin resistance by promoting phosphorylated insulin receptor degradation via increasing late endosomes production
Nature Communications (2023)
-
Co-receptor regulation of insulin signaling
Nature Metabolism (2022)
-
Delving into insulin receptor regulation
Nature Reviews Endocrinology (2022)