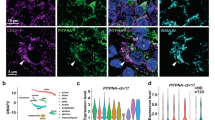
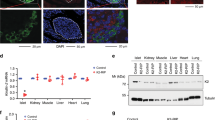
Abstract
MODY8 (maturity-onset diabetes of the young, type 8) is a dominantly inherited monogenic form of diabetes associated with mutations in the carboxyl ester lipase (CEL) gene expressed by pancreatic acinar cells. MODY8 patients develop childhood-onset exocrine pancreas dysfunction followed by diabetes during adulthood. However, it is unclear how CEL mutations cause diabetes. In the present study, we report the transfer of CEL proteins from acinar cells to β-cells as a form of cross-talk between exocrine and endocrine cells. Human β-cells show a relatively higher propensity for internalizing the mutant versus the wild-type CEL protein. After internalization, the mutant protein forms stable intracellular aggregates leading to β-cell secretory dysfunction. Analysis of pancreas sections from a MODY8 patient reveals the presence of CEL protein in the few extant β-cells. The present study provides compelling evidence for the mechanism by which a mutant gene expressed specifically in acinar cells promotes dysfunction and loss of β-cells to cause diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq data in EndoC-βH1 cells have been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus under accession no. GSE185430. Other data that support the findings of the present study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Rickels, M. R., Norris, A. W. & Hull, R. L. A tale of two pancreases: exocrine pathology and endocrine dysfunction. Diabetologia 63, 2030–2039 (2020).
Söling, H. D. & Unger, K. O. The role of insulin in the regulation of α-amylase synthesis in the rat pancreas. Eur. J. Clin. Invest. 2, 199–212 (1972).
Adler, G. & Kern, H. F. Regulation of exocrine pancreatic secretory process by insulin in vivo. Horm. Metab. Res. 7, 290–296 (1975).
Henderson, J. R. Why are the islets of Langerhans? Lancet 294, 469–470 (1969).
Aida, K. et al. Crucial role of Reg I from acinar-like cell cluster touching with islets (ATLANTIS) on mitogenesis of beta cells in EMC virus-induced diabetic mice. Biochem. Biophys. Res. Commun. 503, 963–969 (2018).
Aida, K. et al. Distinct cell clusters touching islet cells induce islet cell replication in association with over-expression of regenerating gene (REG) protein in fulminant type 1 diabetes. PLoS ONE 9, e95110 (2014).
Egozi, A., Bahar Halpern, K., Farack, L., Rotem, H. & Itzkovitz, S. Zonation of pancreatic acinar cells in diabetic mice. Cell Rep. 32, 108043 (2020).
Czakó, L., Hegyi, P., Rakonczay, Z., Wittmann, T. & Otsuki, M. Interactions between the endocrine and exocrine pancreas and their clinical relevance. Pancreatology 9, 351–359 (2009).
Radlinger, B., Ramoser, G. & Kaser, S. Exocrine pancreatic insufficiency in type 1 and type 2 diabetes. Curr. Diab. Rep. 20, 18 (2020).
Sasikala, M. et al. β-Cell dysfunction in chronic pancreatitis. Dig. Dis. Sci. 57, 1764–1772 (2012).
Sheikh, S. et al. Reduced β-cell secretory capacity in pancreatic-insufficient, but not pancreatic-sufficient, cystic fibrosis despite normal glucose tolerance. Diabetes 66, 134–144 (2017).
Andersen, D. K. et al. Diabetes, pancreatogenic diabetes, and pancreatic cancer. Diabetes 66, 1103–1110 (2017).
Raeder, H. et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat. Genet. 38, 54–62 (2006).
Tjora, E. et al. Severe pancreatic dysfunction but compensated nutritional status in monogenic pancreatic disease caused by carboxyl-ester lipase mutations. Pancreas 42, 1078–1084 (2013).
Raeder, H. et al. Carboxyl-ester lipase maturity-onset diabetes of the young is associated with development of pancreatic cysts and upregulated MAPK signaling in secretin-stimulated duodenal fluid. Diabetes 63, 259–269 (2014).
Johansson, B. B. et al. The role of the carboxyl ester lipase (CEL) gene in pancreatic disease. Pancreatology 18, 12–19 (2018).
Lombardo, D. Bile salt-dependent lipase: its pathophysiological implications. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1533, 1–28 (2001).
Kolar, M. J. et al. Branched fatty acid esters of hydroxy fatty acids are preferred substrates of the MODY8 protein carboxyl ester lipase. Biochemistry 55, 4636–4641 (2016).
Johansson, B. B. et al. Diabetes and pancreatic exocrine dysfunction due to mutations in the carboxyl ester lipase gene-maturity onset diabetes of the young (CEL-MODY): a protein misfolding disease. J. Biol. Chem. 286, 34593–34605 (2011).
Torsvik, J. et al. Endocytosis of secreted carboxyl ester lipase in a syndrome of diabetes and pancreatic exocrine dysfunction. J. Biol. Chem. 289, 29097–29111 (2014).
Xiao, X. et al. A carboxyl ester lipase (CEL) mutant causes chronic pancreatitis by forming intracellular aggregates that activate apoptosis. J. Biol. Chem. 291, 23224–23236 (2016).
Dalva, M. et al. Pathogenic carboxyl ester lipase (CEL) variants interact with the normal CEL protein in pancreatic cells. Cells 9, 244 (2020).
Dirice, E. et al. Soluble factors secreted by T cells promote β-cell proliferation. Diabetes 63, 188–202 (2014).
Niederau, C., Fronhoffs, K., Klonowski, H. & S., H. Active pancreatic digestive enzymes show striking differences in their potential to damage isolated rat pancreatic acinar cells. J. Lab. Clin. Med. 125, 265–275 (1995).
Shen, D. et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 60, 173–185 (2011).
Shpetner, H., Joly, M., Hartley, D. & Corvera, S. Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J. Cell Biol. 132, 595–605 (1996).
Trajkovic, K. et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008).
Soto, C. & Pritzkow, S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 21, 1332–1340 (2018).
Riss, T. L. & Moravec, R. A. Use of multiple assay endpoints to investigate the effects of incubation time, dose of toxin, and plating density in cell-based cytotoxicity assays. Assay Drug Dev. Technol. 2, 51–62 (2004).
Cozar-Castellano, I., Haught, M. & Stewart, A. F. The cell cycle inhibitory protein p21cip is not essential for maintaining β-cell cycle arrest or β-cell function in vivo. Diabetes 55, 3271–3278 (2006).
Alcorta, D. A. et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl Acad. Sci. USA 93, 13742–13747 (1996).
Aguayo-Mazzucato, C. et al. Acceleration of β cell aging determines diabetes and senolysis improves disease outcomes. Cell Metab. 30, 129–142.e4 (2019).
Stancill, J. S. et al. Chronic β-cell depolarization impairs β-cell identity by disrupting a network of Ca2+-regulated genes. Diabetes 66, 2175–2187 (2017).
Gregg, T. et al. Pancreatic β-cells from mice offset age-associated mitochondrial deficiency with reduced KATP channel activity. Diabetes 65, 2700–2710 (2016).
Liu, S. et al. Insulin signaling regulates mitochondrial function in pancreatic β-cells. PLoS ONE 4, e7983 (2009).
Abulizi, A. et al. Multi-tissue acceleration of the mitochondrial phosphoenolpyruvate cycle improves whole-body metabolic health. Cell Metab. 32, 751–766 (2020).
Taneera, J. et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum. Mol. Genet. 24, 1945–1955 (2014).
Eizirik, D. L., Miani, M. & Cardozo, A. K. Signalling danger: endoplasmic reticulum stress and the unfolded protein response in pancreatic islet inflammation. Diabetologia 56, 234–241 (2013).
Kahraman, S., Okawa, E. R. & Kulkarni, R. N. Is transforming stem cells to pancreatic beta cells still the holy grail for type 2 diabetes? Curr. Diab. Rep. 16, 70 (2016).
González, F. et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15, 215–226 (2014).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Pagliuca, F. W. et al. Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014).
Rezania, A. et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes 61, 2016–2029 (2012).
Ræder, H. et al. Absence of diabetes and pancreatic exocrine dysfunction in a transgenic model of carboxyl-ester lipase-MODY (maturity-onset diabetes of the young). PLoS ONE 8, e60229 (2013).
de Boer, P. et al. Large-scale electron microscopy database for human type 1 diabetes. Nat. Commun. 11, 2475 (2020).
Michon, L. et al. Involvement of gap junctional communication in secretion. Biochim. Biophys. Acta Biomembr. 1719, 82–101 (2005).
Hayden, M. R. et al. Attenuation of endocrine-exocrine pancreatic communication in type 2 diabetes: pancreatic extracellular matrix ultrastructural abnormalities. J. Cardiometab. Syndr. 3, 234–243 (2008).
Hellman, B., Wallgren, A. & Petersson, B. Cytological characteristics of the exocrine pancreatic cells with regard to their position in relation to the islets of Langerhans. Acta Endocrinol. 39, 465–473 (2008).
Dybala, M. P. et al. Integrated pancreatic blood flow: bidirectional microcirculation between endocrine and exocrine pancreas. Diabetes 69, 1439–1450 (2020).
Shamir, R. et al. Pancreatic carboxyl ester lipase: a circulating enzyme that modifies normal and oxidized lipoproteins in vitro. J. Clin. Invest. 97, 1696–1704 (1996).
Thompson, P. J. et al. Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metab. 29, 1045–1060.e10 (2019).
Mahadevan, N. R. et al. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc. Natl Acad. Sci. USA 108, 6561–6566 (2011).
Pullen, T. J. & Rutter, G. A. When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes Obes. Metab. 15, 503–512 (2013).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Kahraman, S. et al. Harnessing reaction-based probes to preferentially target pancreatic β-cells and β-like cells. Life Sci. Alliance 4, e202000840 (2021).
Teo, A. K. K. et al. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 288, 5353–5356 (2013).
Brown, W. J. Immunoperoxidase methods for localization of antigens in cultured cells and tissues. Curr. Protoc. Cell Biol. https://doi.org/10.1002/0471143030.cb0406s01 (1999).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Kahraman, S. et al. Tracing of islet graft survival by way of in vivo fluorescence imaging. Diabetes Metab. Res. Rev. 27, 575–583 (2011).
Acknowledgements
We thank E. Tjora (University of Bergen) for providing skin biopsies of a MODY8 family, A. B. Goldfine (Joslin) for providing skin biopsies from a non-family control, D. Hoem (Haukeland University Hospital) for providing pancreatic tissue from a MODY8 patient, A. K. K. Teo (Joslin) for establishing control skin fibroblast cultures and expanding fibroblasts, B. Slipp (Joslin) for technical assistance, O. Ijaduola (Joslin) for maintaining NSG mice, H. Pan and J. Dreyfuss (Joslin Bioinformatics & Biostatistics Core) for analysing RNA-seq data, and C. Cahill (Joslin Advanced Microscopy Core) for assistance with confocal microscopy and processing samples for electron microscopy. We thank P. R. Njølstad (University of Bergen) for support and discussions throughout the study and S. Bonner-Weir (Joslin) and M. Solimena (Dresden) for discussions. We thank the IIDP for providing human pancreatic islets (NIH grant no. 2UC4DK098085) and Prodo Labs for providing human pancreatic acinar and islet tissues. Flow cytometry experiments were performed in the Joslin Flow Cytometry Core, supported by the Diabetes Research Center (DRC) (nos. P30DK036836 and S10 OD021740-01). R.N.K. acknowledges support from the NIH (grant nos. R01 DK067536 and R01 DK103215). A.M. acknowledges support from the Western Norway Regional Health Authority (Helse Vest, no. 912057) and the Research Council of Norway (FRIMEDBIO, no. 289534). H.R. acknowledges support from Bergen Forskningsstiftelse (no. BFS2014REK02), Diabetesforbundet, Novo Nordisk Foundation (no. NNF17OC0027258), Johan Selmer Kvanes legat and the Western Norway Regional Health Authority (grant no. 911985), and D.H. acknowledges support from NIH/National Institute of Diabetes and Digestive and Kidney Diseases (grant no. R01DK096239).
Author information
Authors and Affiliations
Contributions
S.K. conceived the idea, designed and performed the experiments, analysed the data and wrote the manuscript. E.D. and G.B. performed transplantation experiments. E.D. contributed to confocal imaging. D.D. contributed to cell culture experiments, western blotting, immunohistochemistry and confocal imaging. M.K.G. performed a Seahorse assay. J.H. contributed to immunohistochemistry. L.H. and S.K.M. contributed to acinar differentiation of hiPSC and hESC lines. C.L.S. and D.H. contributed to generation of isogenic hESC lines. H.R. provided MODY8 patient-derived skin biopsies and contributed to conceptual discussions. B.B.J. established fibroblast cultures from skin biopsies. B.B.J. and A.M. provided CEL plasmids and stable OE HEK293 cell lines, and contributed to conceptual discussions. A.M. and J.A. contributed to immunohistochemistry on MODY8 patient pancreas sections. R.N.K. conceived the idea, contributed to discussions, designed the experiments, supervised the project and wrote the manuscript. All the authors reviewed, commented on and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Guy Rutter and the other, anonymous, reviewers for their contribution to the peer review of this work. The primary handling editor was Isabella Samuelson
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Reduced secretion of mutant CEL protein from 266-6 acinar cells and endocytosis-mediated uptake by β-cells.
a, Mouse acinar cells (266-6) transfected with empty vector (EV), wild-type CEL (WT), or mutant CEL (MUT) plasmids. Cell lysates (L) and medium (M) were collected and labeled with anti-V5 antibody to measure V5-tagged CEL levels. b, Western blots, representative of three independent experiments, show low molecular mass forms of wild-type and mutant human CEL proteins in lysates and the fully glycosylated, high molecular mass forms secreted into media. c-e, The quantification of three biologically independent samples. Fold change relative to EV. Lysate CEL levels were normalized to β-actin. f, Determination of the concentration of CEL protein in conditioned medium by quantitative immunoblotting (n = 2 biologically independent samples) using recombinant mouse CEL proteins (rmCEL). g, A standard curve for rmCEL. h, The concentrations of wild-type and mutant CEL proteins were estimated according to the standard curve. i, Recipient β-cells were treated with conditioned medium obtained from HEK293 donor cells for 30 min at 37 °C or at 4 °C (n = 3 biologically independent samples). j, The quantification of three biologically independent samples. Fold change relative to WT 37 °C. V5-tagged CEL levels were normalized to α-tubulin levels. k, Percentage of CEL + β-cells detected by immunostaining after treatment with Wortmannin to block endocytosis (n = 3 biologically independent samples). l, Western blot quantification of β-cells treated with conditioned medium obtained from HEK293 donor cells treated with DMSO or exosome inhibitor GW4869. Fold change relative to WT DMSO (n = 3 biologically independent samples). m, Conditioned media collected from HEK293 donor cells stably transfected with EV, WT, or MUT plasmids was subjected to protein aggregation assay (n = 9 biologically independent samples). Fluorescence signal generated by Proteostat detection dye was measured. Aggregated lysozyme (20 μg) and monomeric lysozyme (20 μg) were used as positive and negative controls, respectively (n = 3 independent samples). Data are expressed as fold change relative to WT. Data are presented as mean values ± SEM. One-way ANOVA followed by Tukey’s multiple comparison test (c,d,j,k-m), two-tailed t-test used for (e). Dashed line is added for easy comprehension (b,f,i).
Extended Data Fig. 2 Accumulation of mutant CEL protein aggregates reduces proliferation and impairs function of β-cells.
a, Representative FACS plots showing percentage of AnnexinV and Zombie Near Infrared (NIR) stained β-cells treated with conditioned medium for 10 days to assess apoptosis levels (n = 3 biologically independent samples). b-d, Heatmap showing differentially expressed genes involved in cellular senescence (b), Insulin secretion (c), glycolysis/gluconeogenesis (top panel), oxidative phosphorylation (lower panel) (d) in β-cells treated with conditioned medium for 10 days (EV n = 9, WT n = 10, MUT n = 10 biologically independent samples). e, Mitochondrial respiration profile of EndoC-βH1 cells exposed to conditioned media for 10 days. Cells were challenged with oligomycin (15 μM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (10 μM), and Rotenone (1.1 µM) and actinomycin (25 μM) (left panel). Quantification of basal (middle panel) and maximal respiration (right panel) capacity (EV n = 5, WT n = 4, MUT n = 5 biologically independent samples). Data are presented as mean values ± SEM. One-way ANOVA followed by Tukey’s multiple comparison (a,e).
Extended Data Fig. 3 Generation of hESC lines expressing mutant CEL.
a, Schematic of CRISPR-Cas9 strategy for generation of CEL mutant hESC lines. Exons and introns are represented by boxes and lines, respectively. A 100-nt single strand DNA (ssDNA) carrying the patient specific mutation (c.1686delT) and two gRNAs (gRNA1, gRNA2) were used to target the first repeat of VNTR in the CEL gene exon 11. PAM sequences in the gRNAs are indicated in purple. Heterozygous and compound heterozygous mutant cell lines were generated. Deletions are indicated in red boxes. b, Alignment of the C-terminal end of WT and MUT CEL proteins. WT and MUT amino acid sequences are indicated in blue and red, respectively. Small deletions cause frameshift in the first repeat of VNTR (Rep1) and create a shorter protein. Each asterisk denotes deletion of an amino acid. c, DNA sequencing of isogenic hESC lines carrying deletion mutations in the CEL gene. Asterisks show the mutation sites. d, Normal karyotype of WT and MUT hESC lines generated by CRISPR-Cas9 technology.
Extended Data Fig. 4 Generation and characterization of MODY8 disease-specific hiPSCs.
a, MODY8 family pedigree. Solid symbols denote diabetes. NN, no mutation; NM, mutation. b, Outline of the episomal reprogramming approach. Details are given in the Methods section. c, Representative bright-field images of hiPSC colonies derived from family and non-family controls (Fam Ctr n = 4, and Non-Fam Ctr n = 4 independent clones) and from mutation carriers with or without diabetes (Mut + Dia+ n = 8, Mut+ Dia- n = 6 independent clones). Scale bar is 1 mm. d, Normal karyotype of hiPSCs derived from controls and MODY8 patients (Non-Fam Ctr n = 2, Fam Ctr n = 2, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples). e, DNA sequencing confirmed presence of c.1686delT deletion (asterisk) in hiPSCs derived from MODY8 patients (Non-Fam Ctr n = 2, Fam Ctr n = 2, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples). f, Immunostaining for pluripotency markers OCT4 (red), SOX2 (green), SSEA4 (red) in hiPSCs derived from controls and mutation carriers (Non-Fam Ctr n = 2, Fam Ctr n = 2, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples). Nuclei stained with DAPI (blue). Scale bar is 500 μm. g, Control and MODY8 hiPSCs formed teratoma approximately 13 weeks after implantation into immunodeficient mice. Non-Fam Ctr n = 3, Fam Ctr n = 3, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples. Data are presented as mean values ± SEM. The difference between control vs mutant lines is not significant. Two-tailed t-test. h, Control and mutant hiPSCs formed teratoma (approximately 2 cm diameter) after injection into right leg muscle of immunodeficient mice (Non-Fam Ctr n = 3, Fam Ctr n = 3, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples). i, Representative images of teratomas stained with hematoxylin and eosin (HE) (Non-Fam Ctr n = 3, Fam Ctr n = 3, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples). Arrows show differentiated tissues including neural rosettes (ectoderm), cartilage (mesoderm), epithelial tissue (endoderm). Scale bar 50 μm.
Extended Data Fig. 5 Differentiation of control hiPSCs to generate S6 β-like cells.
a, Differentiation protocol used to differentiate hPSCs towards insulin expressing β-like cells. Details are given in Supplementary Table 4. b, Control hiPSCs (family and non-family control) were differentiated to S6 and expression levels of genes specific to β-cells were measured. Two differentiation batches of control hiPSCs (Batch1 n = 4, Batch2 n = 2 biologically independent samples) and human islets collected from five donors were used. Donor information is provided in Supplementary Table 1. β-Actin was used as a housekeeping control. Fold change relative to S0. Data are presented as mean values ± SEM. c, Control hiPSCs (N65-51 family control, n = 2 biologically independent samples) were differentiated to S6 and co-stained for several pancreatic markers; INS/GCG, NKX6.1/PDX1, CHGA/GCG, PDX1/ISL1, NeuroD1/PDX1, NKX2.2/PDX1. Nuclei stained with DAPI (blue). Scale bar is 500 μm.
Extended Data Fig. 6 Differentiation of gene-edited hESCs and patient-derived hiPSCs to definitive endoderm stage.
a, FACS analysis of wild-type and mutant hESC lines differentiated to S1. + / + n = 12, +/1686delT n = 3, +/1698delA n = 3, +/1690_1702del n = 3, +/1683_1704del n = 3, 1686delT/1702delG n = 3 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in wild-type vs. each mutant line by two-tailed multiple t-test corrected using Holm-Sidak method. b, FACS analysis of control and mutant hiPSC lines differentiated to S1. Control n = 8, Mut+ Dia- n = 6, Mut+ Dia+ n = 7 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in control vs Mut+ Dia- and vs. Mut+ Dia+ by two-tailed multiple t-test corrected using Holm-Sidak method. c, Immunostaining images, representative of three biologically independent samples, showing DE cells stained for SOX17 (green) and OCT4 (red). Nuclei stained with DAPI (blue). Scale bar is 500 μm.
Extended Data Fig. 7 Differentiation of gene-edited hESCs and patient-derived hiPSCs to pancreatic progenitor stage.
a, FACS analysis of wild-type and mutant hESC lines differentiated to S4. + / + n = 16, +/1686delT n = 4, +/1698delA n = 4, +/1690_1702del n = 4, +/1683_1704del n = 4, 1686delT/1702delG n = 4 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in wild-type vs. each mutant line by two-tailed multiple t-test corrected using Holm-Sidak method. c, Representative FACS plots of control and mutant (with or without diabetes) hiPSC lines differentiated to S4. b, FACS analysis of control and mutant hiPSC lines differentiated to S4. Control n = 6, Mut+ Dia- n = 5, Mut+ Dia+ n = 4 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in control vs Mut+ Dia- and vs Mut+ Dia+ by two-tailed multiple t-test corrected using Holm-Sidak method. c, Immunostaining images, representative of three biologically independent samples, showing PP cells for PDX1 (green) and NKX6.1 (red). Nuclei stained with DAPI (blue). Scale bar is 500 μm.
Extended Data Fig. 8 Differentiation of gene-edited hESCs and patient-derived hiPSCs to β-like cells.
a, FACS analysis of wild-type and mutant hESC lines differentiated to S6. + / + n = 4, +/1686delT n = 4, +/1698delA n = 3, +/1690_1702del n = 4, +/1683_1704del n = 4, 1686delT/1702delG n = 4 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in wild-type vs. each mutant line by two-tailed multiple t-test corrected using Holm-Sidak method. b, FACS analysis of control and mutant hiPSC lines differentiated to S6. Control n = 8, Mut+ Dia- n = 6, Mut+ Dia+ n = 6 biologically independent samples. Data are presented as mean values ± SEM. No difference was detected in control vs. Mut+ Dia- and vs. Mut+ Dia+ by two-tailed multiple t-test corrected using Holm-Sidak method. c, Immunostaining images, representative of three biologically independent samples, showing β-like cells for CPEP (green) and GCG (red). Nuclei stained with DAPI (blue). Scale bar is 500 μm. d, GSIS was performed by stimulating wild-type or mutant S6 cells with 1 mM low glucose (LG) or 20 mM high glucose (HG) for an hour. The stimulation index was calculated as the fold increase in human C-peptide release measured in 20 mM over 1 mM glucose. +/+ n = 4, +/1686delT n = 4, 1686delT/1702delG n = 4 biologically independent samples. Data are represented as median with 25% to 75% percentile box and min/max whisker plots. Two-tailed multiple t-tests followed by Holm Sidak’s multiple comparison test. e, Stimulation index of iPSC-derived β-like cells. Control n = 4, Mut+ Dia- n = 4, Mut+ Dia+ n = 4 biologically independent samples. Data are represented as median with 25% to 75% percentile box and min/max whisker plots. Two-tailed multiple t-tests followed by Holm Sidak’s multiple comparison test.
Extended Data Fig. 9 Differentiation of gene-edited hESCs and patient-derived hiPSCs to acinar-like organoids.
a, Differentiation protocol used to differentiate hPSCs towards exocrine organoids. Details are given in the Methods section. b, Bright field images, representative of three biologically independent samples, show organoids that were derived from gene-edited hESCs. Scale bar is 100 μm. c, Expression levels of genes specific to exocrine pancreas were measured by RT-PCR (WT n = 5, MUT n = 5 biologically independent samples). Data are presented as mean values ± SEM. No difference was detected in wild-type vs. mutant by two-tailed t-test. Similar results were observed using three differentiation batches of hESC or hiPSC lines. β-Actin was used as a housekeeping control. Fold change relative to S0.
Extended Data Fig. 10 Analysis of WT and MUT S4 graft sections.
Representative immunostaining images of grafts derived from mutant (n = 4) or wild-type (n = 4) S4 cells stained for β-cell markers such as INS in green, PDX1, NKX2.2, NeuroD1, and NKX6.1 in red. Human foetal pancreas (34 weeks, n = 1 donor) was used as control. Donor information is given in Supplementary Table 1. Nuclei stained with DAPI (blue). Scale bar is 20 μm.
Supplementary information
Supplementary Information
Supplementary Fig. 1, Protocols and References.
Supplementary Tables
Supplementary Table 1 Donor information. Supplementary Table 2 Antibody information. Supplementary Table 3 Primer information. Supplementary Table 4 In vitro differentiation protocol.
Source data
Source Data Fig. 1
Unprocessed western blots for Fig. 1.
Source Data Fig. 3
Unprocessed western blots for Fig. 3.
Source Data Fig. 4
Unprocessed western blots for Fig. 4.
Source Data Fig. 6
Unprocessed western blots for Fig. 6.
Source Data Extended Data Figure 1
Unprocessed western blots for Extended Data Fig. 1.
Rights and permissions
About this article
Cite this article
Kahraman, S., Dirice, E., Basile, G. et al. Abnormal exocrine–endocrine cell cross-talk promotes β-cell dysfunction and loss in MODY8. Nat Metab 4, 76–89 (2022). https://doi.org/10.1038/s42255-021-00516-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00516-2
This article is cited by
-
Single-nucleus multi-omics of human stem cell-derived islets identifies deficiencies in lineage specification
Nature Cell Biology (2023)
-
Monogenic diabetes
Nature Reviews Disease Primers (2023)
-
Towards a better understanding of diabetes mellitus using organoid models
Nature Reviews Endocrinology (2023)
-
Clinical and genetic characteristics of CEL-MODY (MODY8): a literature review and screening in Chinese individuals diagnosed with early-onset type 2 diabetes
Endocrine (2023)