Abstract
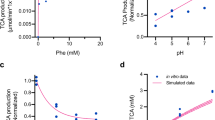
Phenylketonuria (PKU) is a rare disease caused by biallelic mutations in the PAH gene that result in an inability to convert phenylalanine (Phe) to tyrosine, elevated blood Phe levels and severe neurological complications if untreated. Most patients are unable to adhere to the protein-restricted diet, and thus do not achieve target blood Phe levels. We engineered a strain of E. coli Nissle 1917, designated SYNB1618, through insertion of the genes encoding phenylalanine ammonia lyase and l-amino acid deaminase into the genome, which allow for bacterial consumption of Phe within the gastrointestinal tract. SYNB1618 was studied in a phase 1/2a randomized, placebo-controlled, double-blind, multi-centre, in-patient study (NCT03516487) in adult healthy volunteers (n = 56) and patients with PKU and blood Phe level ≥600 mmol l−1 (n = 14). Participants were randomized to receive a single dose of SYNB1618 or placebo (part 1) or up to three times per day for up to 7 days (part 2). The primary outcome of this study was safety and tolerability, and the secondary outcome was microbial kinetics. A D5-Phe tracer (15 mg kg−1) was used to study exploratory pharmacodynamic effects. SYNB1618 was safe and well tolerated with a maximum tolerated dose of 2 × 1011 colony-forming units. Adverse events were mostly gastrointestinal and of mild to moderate severity. All participants cleared the bacteria within 4 days of the last dose. Dose-responsive increases in strain-specific Phe metabolites in plasma (trans-cinnamic acid) and urine (hippuric acid) were observed, providing a proof of mechanism for the potential to use engineered bacteria in the treatment of rare metabolic disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data in the published article (and its Supplementary Information files) have been presented where possible as group-level summaries. Any data presented to illustrate individual patient performance have been de-identified. The data sets generated during and/or analysed during the current study are available from the corresponding author (M.K.P) upon reasonable request including a methodologically sound proposal, although restrictions may apply due to patient privacy and HIPAA regulations.
Change history
16 August 2022
A Correction to this paper has been published: https://doi.org/10.1038/s42255-022-00635-4
References
DeGroot, M. J., Hoeksma, M., Blau, N., Reijngoud, D. J. & Van Spronsen, F. J. Pathogenesis of cognitive dysfunction in phenylketonuria: review of hypotheses. Mol. Genet. Metab. 99, S86–S89 (2010).
Vockley, J. et al. Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet. Med. 16, 188–200 (2014).
Van Spronsen, F. J. et al. Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 5, 743–756 (2017).
Van Wegberg, A. M. J. et al. The complete European guidelines on phenylketonuria: diagnosis and treatment. Orphanet J. Rare Dis. 12, 162 (2017).
Singh, R. H. et al. Recommendations for the nutrition management of phenylalanine hydroxylase deficiency. Genet. Med. 16, 121–131 (2014).
Jurecki, E. R. et al. Adherence to clinic recommendations among patients with phenylketonuria in the United States. Mol. Genet. Metab. 120, 190–197 (2017).
Brown, C. S. & Lichter-Konecki, U. Phenylketonuria (PKU): a problem solved? Mol. Genet. Metab. Rep. 6, 8–12 (2016).
Daelman, L., Sedel, F. & Tourbah, A. Progressive neuropsychiatric manifestations of phenylketonuria in adulthood. Rev. Neurol. 170, 280–287 (2014).
Bilder, D. A. et al. Systematic review and meta-analysis of neuropsychiatric symptoms and executive functioning in adults with phenylketonuria. Dev. Neuropsychol. 41, 245–260 (2016).
FDA. Kuvan prescribing information. BioMarin Pharmaceutical https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/022181s013lbl.pdf (2014).
Hoskins, J. A. The occurrence, metabolism and toxicity of cinnamic acid and related compounds. J. Appl. Toxicol. 4, 283–292 (1984).
Kim, W. et al. Trends in enzyme therapy for phenylketonuria. Mol. Ther. 10, 220–224 (2004).
Lindegren, M. L. et al. A systematic review of BH4 (sapropterin) for the adjuvant treatment of phenylketonuria. JIMD Rep. 8, 109–119 (2013).
Vernon, H. J. et al. Introduction of sapropterin dihydrochloride as standard of care in patients with phenylketonuria. Mol. Genet. Metab. 100, 229–233 (2010).
FDA. Palynziq prescribing information. BioMarin Pharmaceutical https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/761079s000lbl.pdf (2018).
Chang, T. M., Bourget, L. & Lister, C. A new theory of enterorecirculation of amino acids and its use for depleting unwanted amino acids using oral enzyme-artificial cells, as in removing phenylalanine in phenylketonuria. Artif. Cells Blood Substit. Immobil. Biotechnol. 23, 1–21 (1995).
Sarkissian, C. N. et al. A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc. Natl Acad. Sci. USA 96, 2339–2344 (1999).
Levy, H. L., Sarkissian, C. N. & Scriver, C. R. Phenylalanine ammonia lyase (PAL): from discovery to enzyme substitution therapy for phenylketonuria. Mol. Genet. Metab. 24, 223–229 (2018).
Hoskins, J. A. et al. Enzymatic control of phenylalanine intake in phenylketonuria. Lancet 23, 392–394 (1980).
Bourge, L. & Chang, T. M. Effects of oral administration of artificial cells immobilized phenylalanine ammonia-lyase on intestinal amino acids of phenylketonuric rats. Biomater. Artif. Cells Artif. Organs 17, 161–181 (1989).
Fabio Parmeggiani, N., Weise, J., Ahmed, S. T. & Turner, N. J. Synthetic and therapeutic applications of ammonia-lyases and aminomutases. Chem. Rev. 118, 73–118 (2018).
Pereira de Sousa, I., Gourmel, C., Berkovska, O., Burger, M. & Leroux, J.-C. A microparticulate based formulation to protect therapeutic enzymes from proteolytic digestion: phenylalanine ammonia lyase as case study. Nat. Res. Sci. Rep. 10, 3651 (2020).
Isabella, V. M. et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018).
Crook, N. et al. Adaptive strategies of the candidate E. coli Nissle in the mammalian gut. Cell Host Microbe 25, 499–512 (2019).
Kurtz, C. B. et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 16, eaau7975 (2019).
Scriver, C. R. & Rosenberg, L. E. Amino Acid Metabolism and its Disorders (Saunders, 1973).
Ardeypharm. Mutaflor Consumer Information https://www.mutaflor.com.au/wp-content/uploads/2014/05/Consumer-Package-Insert-Mutaflor.pdf (2012).
Bioanalytical Method Validation: Guidance for Industry (US Food and Drug Administration, 2018); https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf
R Core Team. R: a Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020); https://www.R-project.org/
Lenth, R. V. emmeans: Estimated Marginal Means, aka Least-Squares Means. R package v.1.5.0 (2020); https://CRAN.R-project.org/package=emmeans
Acknowledgements
We are grateful to the study patients and their families, and to the research nurses and study staff, as well as the National PKU Association for all their support. We thank N. Longo and C. Harding for their invaluable scientific advice. We greatly appreciate the technical and editorial support provided by J. Blasbalg, C. Anderson, P. Cantarella, M. James, M. Daza and K. Pace. Funding for this study was provided by Synlogic, Inc. The funders participated in study design, data analysis and preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization and design of the work were carried out by M.K.P., W.S.D., L.B., V.M.I., C.B.K. and A.M.B. The acquisition of data was performed by M.K.P., L.B., J.V., S.L.S, S.J.S. and J.A.P. The analysis and interpretation of data were performed by M.K.P., W.S.D., B.D.G., D.A.W., M.J.C., M.R.C., R.J.R., C.B.K. and A.M.B. Trial delivery and administration were carried out by L.B. and V.V.S. The original draft of the manuscript was written by M.K.P., W.S.D., V.V.S., M.J.C., M.R.C., R.J.R., C.B.K. and A.M.B. The submitted paper was reviewed and approved by all the authors.
Corresponding author
Ethics declarations
Competing interests
J.V. reports research funding from Biomarin Pharmaceuticals, Homology Pharmaceuticals, Nestle, PTC Therapeutics, outside the submitted work. M.K.P., L.B., V.V.S., V.M.I., M.J.C., M.R.C., R.J.R., C.B.K. and A.M.B. are full-time employees and hold equity in Synlogic Inc. D.A.W. receives a salary from Metabolic Solutions. Metabolic Solutions was paid a fee to analyse samples for this publication. W.S.D. is a consultant for Synlogic Inc. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Metabolism thanks Nicole Mayer Hamblett and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: George Caputa.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SYNB1618 Mechanism of Action.
SYNB1618 contains chromosomally inserted genes encoding PheP, a high-affinity Phe transporter that can bring Phe into the cell cytoplasm, PAL, which converts Phe to trans-cinnamate, and LAAD, which converts Phe to phenylpyruvate. Induction of these components is carried out partially by the anaerobic-responsive transcriptional activator FNR, for strain activation of PAL and pheP in the anoxic environment of the mammalian gut. Additional copies of PAL and LAAD are placed under control of the Isopropyl β-D-1-thiogalactopyranoside, and L-arabinose inducible promoters, respectively, for strain activation in vitro during production of drug product. Abbreviations: AraC = arabinose-responsive transcriptional regulator; FNR = fumarate and nitrate reductase regulator; LAAD = L-amino acid deaminase from Proteus mirabilis; PAL = phenylalanine ammonia lyase from Photorhabdus luminescens; PheP = high-affinity phenylalanine transporter; pheP = gene encoding high-affinity phenylalanine transporter; Ptac = synthetic promoter controlling PAL expression and regulated by the LacI transcriptional repressor; ΔdapA = deletion of dapA gene leading to diaminopimelate auxotrophy.
Supplementary information
Supplementary Information
Supplementary Tables 1–6
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Puurunen, M.K., Vockley, J., Searle, S.L. et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nat Metab 3, 1125–1132 (2021). https://doi.org/10.1038/s42255-021-00430-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00430-7
This article is cited by
-
Stable expression of HIV-1 MPER extended epitope on the surface of the recombinant probiotic bacteria Escherichia Coli Nissle 1917 using CRISPR/Cas9
Microbial Cell Factories (2024)
-
Alkaptonuria
Nature Reviews Disease Primers (2024)
-
Drinkable in situ-forming tough hydrogels for gastrointestinal therapeutics
Nature Materials (2024)
-
Pharmacovigilance for rare diseases: a bibliometrics and knowledge-map analysis based on web of science
Orphanet Journal of Rare Diseases (2023)
-
Bacterial therapies at the interface of synthetic biology and nanomedicine
Nature Reviews Bioengineering (2023)