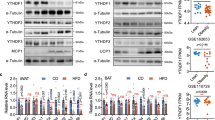
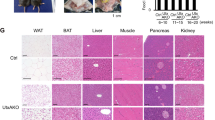
Abstract
Males and females exhibit striking differences in the prevalence of metabolic traits including hepatic steatosis, a key driver of cardiometabolic morbidity and mortality. RNA methylation is a widespread regulatory mechanism of transcript turnover. Here, we show that presence of the RNA modification N6-methyladenosine (m6A) triages lipogenic transcripts for degradation and guards against hepatic triglyceride accumulation. In male but not female mice, this protective checkpoint stalls under lipid-rich conditions. Loss of m6A control in male livers increases hepatic triglyceride stores, leading to a more ‘feminized’ hepatic lipid composition. Crucially, liver-specific deletion of the m6A complex protein Mettl14 from male and female mice significantly diminishes sex-specific differences in steatosis. We further surmise that the m6A installing machinery is subject to transcriptional control by the sex-responsive BCL6–STAT5 axis in response to dietary conditions. These data show that m6A is essential for precise and synchronized control of lipogenic enzyme activity and provide insights into the molecular basis for the existence of sex-specific differences in hepatic lipid traits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq, m6A-seq and ATAC–seq data can be accessed at the NIH Sequence Read Archive repository under accessions PRJNA663718 and GSE157907. Source data are provided with this paper.
Change history
22 February 2023
A Correction to this paper has been published: https://doi.org/10.1038/s42255-023-00748-4
18 July 2022
A Correction to this paper has been published: https://doi.org/10.1038/s42255-022-00614-9
References
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investi 109, 1125–1131 (2002).
Shimomura, I. et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl Acad. Sci. USA 96, 13656–13661 (1999).
Shimano, H. et al. Isoform 1c of sterol regulatory element-binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 99, 846–854 (1997).
Lee, K. N., Pariza, M. W. & Ntambi, J. M. Differential expression of hepatic stearoyl-CoA desaturase gene 1 in male and female mice. Biochim. Biophys. Acta 1304, 85–88 (1996).
Horton, J. D., Shimano, H., Hamilton, R. L., Brown, M. S. & Goldstein, J. L. Disruption of LDL receptor gene in transgenic SREBP-1a mice unmasks hyperlipidemia resulting from production of lipid-rich VLDL. J. Clin. Invest. 103, 1067–1076 (1999).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Meyer, K. D. & Jaffrey, S. R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014).
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 (2018).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205 (2018).
Liu, J. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Sun, Z. et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942 (2012).
Lu, M. et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 18, 388–395 (2012).
Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019).
Zaccara, S. & Jaffrey, S. R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1582–1595 (2020).
Attie, A. D. et al. Relationship between stearoyl-CoA desaturase activity and plasma triglycerides in human and mouse hypertriglyceridemia. J. Lipid Res. 43, 1899–1907 (2002).
Ou, J. et al. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc. Natl Acad. Sci. USA 98, 6027–6032 (2001).
Dentin, R. et al. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Invest. 115, 2843–2854 (2005).
Matsuda, M. et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15, 1206–1216 (2001).
Shao, W., Machamer, C. E. & Espenshade, P. J. Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. J. Lipid Res. 57, 1564–1573 (2016).
Xiong, X. et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660 (2019).
Kleiner, D. E. et al. Design and validation of a histological scoring system for non-alcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Wilson, C., Chen, P. J., Miao, Z. & Liu, D. R. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 38, 1431–1440 (2020).
Liu, X. M., Zhou, J., Mao, Y., Ji, Q. & Qian, S. B. Programmable RNA N6-methyladenosine editing by CRISPR–Cas9 conjugates. Nat. Chem. Biol. 15, 865–871 (2019).
Link, J. C. et al. X chromosome dosage of histone demethylase KDM5C determines sex differences in adiposity. J. Clin. Invest. 130, 5688–5702 (2020).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Sommars, M. A. et al. Dynamic repression by BCL6 controls the genome-wide liver response to fasting and steatosis. eLife 8, e43922 (2019).
Senagolage, M. D. et al. Loss of transcriptional repression by BCL6 confers insulin sensitivity in the setting of obesity. Cell Rep. 25, 3283–3298 (2018).
Kutyavin, V. I. & Chawla, A. BCL6 regulates brown adipocyte dormancy to maintain thermogenic reserve and fitness. Proc. Natl Acad. Sci. USA 116, 17071–17080 (2019).
Zhang, Y., Laz, E. V. & Waxman, D. J. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 32, 880–896 (2012).
Connerney, J., Lau-Corona, D., Rampersaud, A. & Waxman, D. J. Activation of male liver chromatin accessibility and STAT5-dependent gene transcription by plasma growth hormone pulses. Endocrinology 158, 1386–1405 (2017).
Qasem, R. J. et al. Decreased liver triglyceride content in adult rats exposed to protein restriction during gestation and lactation: role of hepatic triglyceride utilization. Clin. Exp. Pharmacol. Physiol. 42, 380–388 (2015).
Zhang, W. et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281, 10105–10117 (2006).
Link, J. C. & Reue, K. Genetic basis for sex differences in obesity and lipid metabolism. Annu Rev. Nutr. 37, 225–245 (2017).
Rinn, J. L. & Snyder, M. Sexual dimorphism in mammalian gene expression. Trends Genet. 21, 298–305 (2005).
Wang, B. & Tontonoz, P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 14, 452–463 (2018).
Lonardo, A. et al. Sex differences in non-alcoholic fatty liver disease: state of the art and identification of research gaps. Hepatology 70, 1457–1469 (2019).
McIntyre, A. B. R. et al. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 10, 6590 (2020).
Xu, J., Teran-Garcia, M., Park, J. H., Nakamura, M. T. & Clarke, S. D. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J. Biol. Chem. 276, 9800–9807 (2001).
Sessler, A. M., Kaur, N., Palta, J. P. & Ntambi, J. M. Regulation of stearoyl-CoA desaturase 1 mRNA stability by polyunsaturated fatty acids in 3T3-L1 adipocytes. J. Biol. Chem. 271, 29854–29858 (1996).
Wang, X. et al. Hepatocyte TAZ/WWTR1 promotes inflammation and fibrosis in non-alcoholic steatohepatitis. Cell Metab. 24, 848–862 (2016).
Rong, X. et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 18, 685–697 (2013).
Sallam, T. et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016).
Zhang, L. et al. Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. eLife 6, e28766 (2017).
Sallam, T. et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 24, 304–312 (2018).
Zhang, Z. et al. Collaborative interactions of heterogenous ribonucleoproteins contribute to transcriptional regulation of sterol metabolism in mice. Nat. Commun. 11, 984 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Casero, D. et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 16, 1282–1291 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Si, Y., Liu, P., Li, P. & Brutnell, T. P. Model-based clustering for RNA-seq data. Bioinformatics 30, 197–205 (2013).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Salmon-Divon, M., Dvinge, H., Tammoja, K. & Bertone, P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11, 415 (2010).
Bailey, T. L. & Machanick, P. Inferring direct DNA binding from ChIP–seq. Nucleic Acids Res. 40, e128 (2012).
Rogers, G. W. et al. High-throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE 6, e21746 (2011).
Kedersha, N. & Anderson, P. Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61–81 (2007).
Acknowledgements
This work was supported by National Institute of Health (NIH) grants (DK118086 to T.S., HL139549 to T.S., HL149766 to T.S., NS111631 to S.R.J., CA186702 to S.R.J., CA236399 to J.C., CA214965 to J.C. and CA243386 to J.C.), an American Heart Association Transformational Project grant (to T.S.) and a Burroughs Wellcome Fund Career Award for Medical Scientists (to T.S.). J.C. is a Leukemia & Lymphoma Society (LLS) Scholar. Schematics for Fig. 1a were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
T.S. conceived and supervised the study. T.S. and D.A.S. designed the study and guided the interpretation of the results and preparation of the manuscript. D.A.S., X.W., J.K. and D.W. performed the majority of experiments and data analysis. Z.Z. performed the ATAC–seq experiments and data analysis. L.V. and K.R. performed the fatty acid oxidation studies and studies determining the influence of chromosomal versus gonadal factors in Mettl14 regulation and guided the interpretation of results. K.J.W. performed lipidomics studies and analysis. A.H.M. and S.R.J. performed m6A measurement via mass spectrometry. A.H.-V. analysed the data for MOBES. J.C. assisted with m6A loss-of-function in vivo studies and cellular knockdown. D.C. performed m6A and RNA-seq analysis. T.S. wrote the manuscript with input from all authors. All authors discussed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.C. is a scientific founder of Genovel Biotech and holds equities with the company, and is also a Scientific Advisor for Race Oncology. A patent related to this work is issued to T.S. The other authors declare no competing interests.
Additional information
Peer review information Nature Metabolism thanks Fabienne Foufelle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Effect of sex and diet composition on lipid metabolism.
a, Fat mass measured by MRI for male and female mice in 2-week cohort (n = 5 per group). b, Body weight of male and female mice in 2-week cohort (n = 5 per group). c, Quantification of serum cholesterol levels of male and female mice in 2-week cohort (n = 5 per group). d, Quantification of total serum triglycerides of male and female mice in 2-week cohort (n = 5 per group). e, Fat mass measured by MRI for male and female mice in 4-week cohort (n = 5 per group). f, Body weight of male and female mice in 4-week cohort (n = 5 per group). g, Quantification of serum cholesterol levels of male and female mice in 4-week cohort (n = 5 per group). h, Quantification of total serum triglycerides of male and female mice in 4-week cohort (n = 5 per group). i, Quantification of total serum NEFA from male and female mice in 4-week cohort (n = 5 per group). Mice were fed indicated diet for 4 weeks and fasted for 4-hrs. prior to sacrifice. Values are mean ± s.e.m. of 5 independent biological replicates (a-i). P values were calculated using one-way analysis of variance (ANOVA) with multi-group comparison (Fisher’s) in a-i. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. The precise n, P values, and details of the statistical testing are provided in the source data file.
Extended Data Fig. 2 Lipogenic genes are highly enriched for m6A under chow diet.
a, Lipidomics PCA plot for 4-week cohort (n = 5 per group). b, Quantification of major lipid species identified in lipidomics analysis of male and female mouse liver for 2-week cohort (n = 5 per group). c, Lipidomics heatmap for 4-week cohort (n = 5 per group). d, Quantification of major lipid species identified in lipidomics analysis of mouse liver for 4-week cohort (n = 5 per group). e, Comparison of number of differentially expressed genes (>2-fold) in WD or HF versus chow-fed male livers determined by RNA-seq and m6A-seq. f, Nucleotide sequences containing the m6A motifs on lipogenic transcripts and relative position of each motif on full-length mRNA. g, Rank order table of genes with greatest fold-change in m6A in male livers (Chow vs. Western diet). h, UCSC browser screenshot showing changes in m6A enrichment for DGAT2 in chow and HF diet-fed male livers. Mice were fed indicated diet for 4 weeks and fasted for 4-hrs. prior to sacrifice. Values are mean ± s.e.m. of 5 independent biological replicates (b,d). P values were calculated using one-way analysis of variance (ANOVA) with multi-group comparison (Fisher’s) in b and d. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. The precise n, P values, and details of the statistical testing are provided in the source data file.
Extended Data Fig. 3 Loss of the m6A methylase METTL14 increases lipogenesis and hepatic triglyceride accumulation.
a, Western blot of Mettl14 in other tissues from WT and Mettl14 L-KO chow-fed males. Equal amounts of protein were pooled from four animals per group and run in triplicate. b, Lipidomics heatmap comparing hepatic lipidome of WT versus Mettl14 L-KO chow-fed males. (n = 8 per group). c, Quantification of various lipid species from lipidomics analysis of chow-fed male livers (n = 8). Statistical analysis was performed using unpaired two-tailed t-test. Values are mean ± SEM. d, Quantification of total food intake for chow-fed male WT and Mettl14 L-KO mice (n = 8 per group). e, Body weight and percent fat measured by MRI for chow-fed male WT and Mettl14 L-KO mice (n = 8 WT mice, n = 7 L-KO mice). f, Quantification of cellular respiratory rate in livers of chow-fed WT and Mettl14 L-KO mice using NADH as the acceptor (n = 5 per group). g, qPCR analysis of fatty acid oxidation gene expression from chow-fed WT and Mettl14 L-KO livers (n = 5). The experiment was repeated two times with similar results. h, Quantification of serum triglyceride levels from chow-fed male WT and Mettl14 L-KO mice (n = 8 per group). i, qPCR analysis of gene expression from liver of WT and Mettl14 L-KO mice (n = 4 per group). The experiment was repeated two times with similar results. j, Western blot of lipogenic protein levels in a second independent cohort of chow-fed male mice. Equal amounts of protein were pooled from eight animals per group and run in triplicate. The experiment was repeated three times with similar results. k, Western blot comparing levels of lipogenic proteins in the setting of chronic Mettl14 deficiency (albumin-cre transgenics or control). Equal amounts of protein were pooled from five animals per group and run in triplicate. The experiment was repeated two times with similar results. l, Quantification of hepatic triglyceride concentrations from livers of albumin-cre transgenic or control mice (n = 4 Cre-N mice, n = 5 Cre-P mice). m, Quantification of lipogenic transcript m6A abundance in WT and Mettl14 L-KO chow-fed livers measured by m6A-IP-qPCR analysis (n = 4). The relative enrichment of m6A in each sample was determined by normalizing to ten-fold input. The experiment was repeated two times with similar results. n, Quantification of lipogenic transcript m6A abundance in WT and Mettl14 L-KO chow-fed livers measured by m6A-IP-qPCR analysis (n = 4). The relative enrichment of m6A in each sample was determined by normalizing to GAPDH. The experiment was repeated two times with similar results. Mice were fed chow diet for 4 weeks and fasted for 4-hrs. prior to sacrifice. Values are mean ± s.e.m. of 4 (I,l-n), 5 (f,g), 6 (d), or 8 (c,e,h) independent biological replicates. P values were calculated using unpaired two-tailed t-test. (c-i,l-n). *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. The precise n, P values, and details of the statistical testing are provided in the source data file.
Extended Data Fig. 4 m6A regulates the stability and cytoplasmic distribution of lipogenic transcripts.
a, qPCR analysis of lipogenic gene expression in livers of WT and Mettl14 L-KO chow-fed male mice (n = 6 per group). The experiment was repeated three times with similar results. b, qPCR analysis of lipogenic pre-mRNA expression using primers that amplify intronic regions from livers of WT and Mettl14 L-KO chow-fed male mice (n = 4 per group). The experiment was repeated three times with similar results. c, Western blot of SREBP-1c from nuclear fraction (mature SREBP-1 [mSREBP-1c]). mSREBP-1c levels in livers of re-fed (4 hr.) mice are shown as reference. Equal amounts of protein were pooled from five animals per group and run in triplicate. The experiment was repeated two times with similar results d, qPCR analysis of lipogenic transcript levels in WT or Mettl14 L-KO livers at 72 hrs. after Mettl14 deletion (n = 4). e, Polysome profiling curve depicting three major cytoplasmic mRNA pools. f, qPCR analysis quantifying amount of lipogenic mRNAs in ribosome-bound fractions in livers of Mettl14 L-KO chow-fed males compared to WT (n = 3 per group). g, mRNA stability assay for SCD1 mRNA in primary hepatocytes harvested from WT and Mettl14 L-KO chow-fed male mice (n = 3 per group). h, qPCR analysis of Mettl14 expression in AML cells used for single-molecule RNA FISH. (n = 5 per group). i, Quantification of hepatic triglyceride content in WD-fed male mice injected with AAV_m6A writers (Mettl14 + Mettl3 + WTAP) or AAV_null (normalized to liver weight) (n = 6 per group). Mice were fed indicated diet for 4 weeks and fasted for 4-hrs. prior to sacrifice except for mice in (i). Mice in (i) were fed NASH diet for 8-weeks and fasted 4-hrs. prior to sacrifice. Values are mean ± s.e.m. of 3 (f,g), 4 (b,d), 5 (h), or 6 (a,i) independent biological replicates. P values were calculated using unpaired two-tailed t-test. (a,b,d,h,i). *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. The precise n, P values, and details of the statistical testing are provided in the source data file.
Extended Data Fig. 5 Regulation of Mettl14 in response to dietary conditions.
a, Quantification of major lipid species identified in lipidomics analysis of male WD-fed WT and Mettl14 L-KO livers (n = 7 WT mice, n = 8 KO mice). b, UCSC browser screenshots showing decreased H3K27ac at Mettl14 promoter under HF diet feeding compared to chow. Mice were fed either standard chow diet (Prolab Isopro RMH 3000, Purina) for 24 weeks or 8 weeks of standard chow diet followed by 16 weeks of HFD (Soltis et al. 2017. Cell Reports). c, UCSC browser screenshots showing regulation of METTL14 promoter by BCL6 and STAT5a in human (Steube et. al 2017. Nat Comm; Gertz et. al 2013. Mol. Cell; respectively). d, UCSC browser screenshots showing ATAC-seq data for Mettl14 promoter region from male mouse liver under different diets and regulation of Mettl14 promoter by BCL6 and STAT5a in male and female mice fed WD (Zhang et. al 2011. Mol Cel Biol). e, qPCR analysis showing fast/re-feed regulation of Bcl6 expression in livers of chow-fed males (n = 5 per group). The experiment was repeated twice with similar results. f, Quantification of m6A abundance on lipogenic transcripts in fasted (4 hr) or re-fed (fasted overnight and then re-fed 4 hr) male livers as measured by m6A-IP-qPCR (n = 4). The experiment was repeated twice with similar results. g, Expression of Mettl3, WTAP in re-fed mice (fasted overnight and then re-fed 4 hr) compared to fasted (4 hr) mice as measured by qPCR analysis (n = 5). h, qPCR analysis of Mettl3 expression in male and female liver under various diets (n = 4 chow-fed females, n = 5 all other groups). Mice were fed the indicated diet for 4 weeks and fasted (4 hr) or re-fed (fasted overnight and then refed for 4-hrs. prior to sacrifice). Values are mean ± s.e.m. of 4 (f), 5 (e,g,h) or 7 (a) indpendent biological replicates. P values were calculated using unpaired two-tailed t-test (a,e-g) or one-way analysis of variance (ANOVA) followed by multi-group comparison (Fisher’s) in h. *P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001. The precise n, P values, and details of the statistical testing are provided in the source data file.
Extended Data Fig. 6 Loss of Mettl14 is associated with sex-biased gene expression.
a, Comparison of gene expression fold changes between control and Mettl14 L-KO mice obtained from male (x axis) or female (y axis) samples. b, Comparison of gene expression fold changes between male and female mice obtained from control (x axis) or Mettl14 L-KO (y axis) samples. The linear fit of all fold changes (yellow) has a smaller slope than the Control=KO line (black), highlighting that male/female differences are higher in controls for most genes. c, Hierarchical clustering of samples harvested from liver based on pair-wise distances. Shown is the tree based on the Euclidean distance between all samples (n = 7 WT male mice, n = 8 mice all other groups) based on genome-wide mRNA abundance distributions. d, Expression of XIST and SRY in livers of WT and Mettl14 L-KO mice as measured by qPCR (n = 4). e, Heatmaps of selected differential genes with distinct responses to Mettl14 L-KO. Three specific clusters of genes with sex-specific response to Mettl14 are shown. Highlighted are the name of genes known to be involved in metabolic pathways. Gene names are ordered as in the heatmap. All mice were fed WD for 4 weeks and fasted for 4 hrs. prior to sacrifice. P values were calculated using unpaired two-tailed t-test (d). The precise n, P values, and details of the statistical testing are provided in the source data file.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salisbury, D.A., Casero, D., Zhang, Z. et al. Transcriptional regulation of N6-methyladenosine orchestrates sex-dimorphic metabolic traits. Nat Metab 3, 940–953 (2021). https://doi.org/10.1038/s42255-021-00427-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00427-2
This article is cited by
-
Sex differences in energy metabolism: natural selection, mechanisms and consequences
Nature Reviews Nephrology (2024)
-
m6A modification-tuned sphingolipid metabolism regulates postnatal liver development in male mice
Nature Metabolism (2023)
-
N6-methyladenosine modification governs liver glycogenesis by stabilizing the glycogen synthase 2 mRNA
Nature Communications (2022)