Abstract
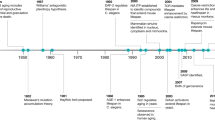
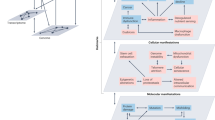
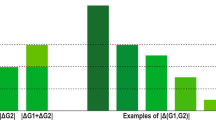
Ageing is the greatest risk factor for most common chronic human diseases, and it therefore is a logical target for developing interventions to prevent, mitigate or reverse multiple age-related morbidities. Over the past two decades, genetic and pharmacologic interventions targeting conserved pathways of growth and metabolism have consistently led to substantial extension of the lifespan and healthspan in model organisms as diverse as nematodes, flies and mice. Recent genetic analysis of long-lived individuals is revealing common and rare variants enriched in these same conserved pathways that significantly correlate with longevity. In this Perspective, we summarize recent insights into the genetics of extreme human longevity and propose the use of this rare phenotype to identify genetic variants as molecular targets for gaining insight into the physiology of healthy ageing and the development of new therapies to extend the human healthspan.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
World Population Ageing 2017 (United Nations, 2017).
Partridge, L., Deelen, J. & Slagboom, P. E. Facing up to the global challenges of ageing. Nature 561, 45–56 (2018).
Barnett, K. et al. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet 380, 37–43 (2012).
Marengoni, A. et al. Aging with multimorbidity: a systematic review of the literature. Ageing Res. Rev. 10, 430–439 (2011).
Goldman, D. The economic promise of delayed aging. Cold Spring Harb. Perspect. Med. 6, a025072 (2015).
Fontana, L., Partridge, L. & Longo, V. D. Extending healthy life span: from yeast to humans. Science 328, 321–326 (2010).
van der Spoel, E. et al. Association analysis of insulin-like growth factor-1 axis parameters with survival and functional status in nonagenarians of the Leiden Longevity Study. Aging (Albany N.Y.) 7, 956–963 (2015).
Passtoors, W. M. et al. Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell 12, 24–31 (2013).
Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993).
Fontana, L. & Partridge, L. Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118 (2015).
Vellai, T. et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620 (2003).
Kapahi, P. et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885–890 (2004).
Johnson, S. C. et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013).
Zhang, Q. et al. Systems-level analysis of human aging genes shed new light on mechanisms of aging. Hum. Mol. Genet. 25, 2934–2947 (2016).
Johnson, S. C., Rabinovitch, P. S. & Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Wilkinson, J. E. et al. Rapamycin slows aging in mice. Aging Cell 11, 675–682 (2012).
Bjedov, I. et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 (2010).
Powers, R. W. III, Kaeberlein, M., Caldwell, S. D., Kennedy, B. K. & Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184 (2006).
Robida-Stubbs, S. et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724 (2012).
Miller, R. A. et al. An Aging Interventions Testing Program: study design and interim report. Aging Cell 6, 565–575 (2007).
Nadon, N. L. et al. Design of aging intervention studies: the NIA interventions testing program. Age (Dordr.) 30, 187–199 (2008).
Harrison, D. E. et al. Acarbose, 17-α-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell 13, 273–282 (2014).
Barzilai, N., Crandall, J. P., Kritchevsky, S. B. & Espeland, M. A. Metformin as a tool to target aging. Cell Metab. 23, 1060–1065 (2016).
Justice, J. N. et al. Development of clinical trials to extend healthy lifespan. Cardiovasc Endocrinol Metab 7, 80–83 (2018).
de Magalhães, J. P. Why genes extending lifespan in model organisms have not been consistently associated with human longevity and what it means to translation research. Cell Cycle 13, 2671–2673 (2014).
Johnson, S. C., Dong, X., Vijg, J. & Suh, Y. Genetic evidence for common pathways in human age-related diseases. Aging Cell 14, 809–817 (2015).
Perls, T. T., Bubrick, E., Wager, C. G., Vijg, J. & Kruglyak, L. Siblings of centenarians live longer. Lancet 351, 1560 (1998).
Schoenmaker, M. et al. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur. J. Hum. Genet. 14, 79–84 (2006).
Herskind, A. M. et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum. Genet. 97, 319–323 (1996).
Christensen, K., Johnson, T. E. & Vaupel, J. W. The quest for genetic determinants of human longevity: challenges and insights. Nat. Rev. Genet. 7, 436–448 (2006).
Murabito, J. M., Yuan, R. & Lunetta, K. L. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J. Gerontol. A Biol. Sci. Med. Sci. 67, 470–479 (2012).
Perls, T. T. et al. Life-long sustained mortality advantage of siblings of centenarians. Proc. Natl Acad. Sci. USA 99, 8442–8447 (2002).
Robine, J. M. & Allard, M. The oldest human. Science 279, 1834–1835 (1998).
Gögele, M. et al. Heritability analysis of life span in a semi-isolated population followed across four centuries reveals the presence of pleiotropy between life span and reproduction. J. Gerontol. A Biol. Sci. Med. Sci. 66, 26–37 (2011).
van den Berg, N. et al. Longevity defined as top 10% survivors and beyond is transmitted as a quantitative genetic trait. Nat. Commun. 10, 35 (2019).
Barzilai, N., Gabriely, I., Gabriely, M., Iankowitz, N. & Sorkin, J. D. Offspring of centenarians have a favorable lipid profile. J. Am. Geriatr. Soc. 49, 76–79 (2001).
Newman, A. B. et al. Health and function of participants in the Long Life Family Study: a comparison with other cohorts. Aging (Albany N.Y.) 3, 63–76 (2011).
Deelen, J. et al. Employing biomarkers of healthy ageing for leveraging genetic studies into human longevity. Exp. Gerontol. 82, 166–174 (2016).
Ash, A. S. et al. Are members of long-lived families healthier than their equally long-lived peers? Evidence from the Long Life Family Study. J. Gerontol. A Biol. Sci. Med. Sci. 70, 971–976 (2015).
Ruby, J. G. et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics 210, 1109–1124 (2018).
Jarry, V., Gagnon, A. & Bourbeau, R. Survival advantage of siblings and spouses of centenarians in 20th-century Quebec. Can. Stud. Popul. 39, 67–78 (2012).
van den Berg, N. et al. Longevity Relatives Count score identifies heritable longevity carriers and suggests case improvement in genetic studies. Aging Cell 19, e13139 (2020).
McDaid, A. F. et al. Bayesian association scan reveals loci associated with human lifespan and linked biomarkers. Nat. Commun. 8, 15842 (2017).
Zenin, A. et al. Identification of 12 genetic loci associated with human healthspan. Commun. Biol. 2, 41 (2019).
Fernandes, M. et al. Systematic analysis of the gerontome reveals links between aging and age-related diseases. Hum. Mol. Genet. 25, 4804–4818 (2016).
Shindyapina, A. V. et al. Germline burden of rare damaging variants negatively affects human healthspan and lifespan. eLife 9, e53449 (2020).
Cash, T. P. et al. Exome sequencing of three cases of familial exceptional longevity. Aging Cell 13, 1087–1090 (2014).
Nygaard, H. B. et al. Whole exome sequencing of an exceptional longevity cohort. J. Gerontol. A Biol. Sci. Med. Sci. 74, 1386–1390 (2019).
Han, J. et al. Discovery of novel non-synonymous SNP variants in 988 candidate genes from 6 centenarians by target capture and next-generation sequencing. Mech. Ageing Dev. 134, 478–485 (2013).
Howden, L. M. & Meyer, J. A. Age and Sex Composition: 2010 (U.S. Census Bureau, 2011).
Andersen, S. L., Sebastiani, P., Dworkis, D. A., Feldman, L. & Perls, T. T. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J. Gerontol. A Biol. Sci. Med. Sci. 67, 395–405 (2012).
Ismail, K. et al. Compression of morbidity is observed across cohorts with exceptional longevity. J. Am. Geriatr. Soc. 64, 1583–1591 (2016).
Sebastiani, P. et al. Families enriched for exceptional longevity also have increased health-span: findings from the Long Life Family Study. Front. Public Health 1, 38 (2013).
Hazra, N. C., Rudisill, C. & Gulliford, M. C. Determinants of health care costs in the senior elderly: age, comorbidity, impairment, or proximity to death? Eur. J. Health Econ. 19, 831–842 (2018).
Abifadel, M. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003).
Zhao, Z. et al. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79, 514–523 (2006).
Hooper, A. J., Marais, A. D., Tanyanyiwa, D. M. & Burnett, J. R. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis 193, 445–448 (2007).
Fitzgerald, K. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383, 60–68 (2014).
Ridker, P. M. et al. Cardiovascular efficacy and safety of bococizumab in high-risk patients. N. Engl. J. Med. 376, 1527–1539 (2017).
Tall, A. R. & Rader, D. J. Trials and tribulations of CETP inhibitors. Circ. Res. 122, 106–112 (2018).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Gorlov, I. P., Gorlova, O. Y., Sunyaev, S. R., Spitz, M. R. & Amos, C. I. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 82, 100–112 (2008).
Kosmicki, J. A., Churchhouse, C. L., Rivas, M. A. & Neale, B. M. Discovery of rare variants for complex phenotypes. Hum. Genet. 135, 625–634 (2016).
Nicolae, D. L. Association tests for rare variants. Annu. Rev. Genomics Hum. Genet. 17, 117–130 (2016).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, eaag1166 (2017).
Oprea, T. I. et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 17, 317–332 (2018).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Tazearslan, C., Huang, J., Barzilai, N. & Suh, Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell 10, 551–554 (2011).
Suh, Y. et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl Acad. Sci. USA 105, 3438–3442 (2008).
Mao, K. et al. Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat. Commun. 9, 2394 (2018).
Vidal, M., Cusick, M. E. & Barabasi, A. L. Interactome networks and human disease. Cell 144, 986–998 (2011).
Barabasi, A. L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011).
Pawson, T. & Nash, P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 14, 1027–1047 (2000).
Wang, X. et al. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol. 30, 159–164 (2012).
Stenson, P. D. et al. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 21, 577–581 (2003).
Khurana, E. et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587 (2013).
Guo, Y. et al. Dissecting disease inheritance modes in a three-dimensional protein network challenges the “guilt-by-association” principle. Am. J. Hum. Genet. 93, 78–89 (2013).
Wei, X. et al. A massively parallel pipeline to clone DNA variants and examine molecular phenotypes of human disease mutations. PLoS Genet. 10, e1004819 (2014).
Chen, S. et al. An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat. Genet. 50, 1032–1040 (2018).
Braun, P. et al. An experimentally derived confidence score for binary protein-protein interactions. Nat. Meth. 6, 91–97 (2009).
Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008).
Yu, H. et al. Next-generation sequencing to generate interactome datasets. Nat. Methods 8, 478–480 (2011).
Sahni, N. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015).
Zhong, Q. et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 5, 321 (2009).
Fabrizio, P., Pozza, F., Pletcher, S. D., Gendron, C. M. & Longo, V. D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 (2001).
Wang, H. D., Kazemi-Esfarjani, P. & Benzer, S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc. Natl Acad. Sci. USA 101, 12610–12615 (2004).
Muñoz, M. J. & Riddle, D. L. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163, 171–180 (2003).
de Magalhães, J. P. & Toussaint, O. GenAge: a genomic and proteomic network map of human ageing. FEBS Lett. 571, 243–247 (2004).
Soldner, F. & Jaenisch, R. Stem cells, genome editing, and the path to translational medicine. Cell 175, 615–632 (2018).
Lo Sardo, V. et al. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell 175, 1796–1810.e1720 (2018).
Aguiar-Oliveira, M. H. & Bartke, A. Growth hormone deficiency: health and longevity. Endocr. Rev. 40, 575–601 (2019).
Tilstra, J. S. et al. NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J. Clin. Invest. 122, 2601–2612 (2012).
Niedernhofer, L. J. et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043 (2006).
Hambright, W. S., Niedernhofer, L. J., Huard, J. & Robbins, P. D. Murine models of accelerated aging and musculoskeletal disease. Bone 125, 122–127 (2019).
Willcox, B. J. et al. FOXO3A genotype is strongly associated with human longevity. Proc. Natl Acad. Sci. USA 105, 13987–13992 (2008).
Morris, B. J., Willcox, D. C., Donlon, T. A. & Willcox, B. J. FOXO3: a major gene for human longevity: a mini-review. Gerontology 61, 515–525 (2015).
Cautain, B. et al. Discovery of a novel, isothiazolonaphthoquinone-based small molecule activator of FOXO nuclear-cytoplasmic shuttling. PLoS ONE 11, e0167491 (2016).
Belguise, K., Guo, S. & Sonenshein, G. E. Activation of FOXO3a by the green tea polyphenol epigallocatechin-3-gallate induces estrogen receptor alpha expression reversing invasive phenotype of breast cancer cells. Cancer Res. 67, 5763–5770 (2007).
Cho, S. et al. Syringaresinol protects against hypoxia/reoxygenation-induced cardiomyocytes injury and death by destabilization of HIF-1α in a FOXO3-dependent mechanism. Oncotarget 6, 43–55 (2015).
Bowman, L. et al. Effects of anacetrapib in patients with atherosclerotic vascular disease. N. Engl. J. Med. 377, 1217–1227 (2017).
Hall, S. S. Genetics: a gene of rare effect. Nature 496, 152–155 (2013).
Rajpathak, S. N. et al. Lifestyle factors of people with exceptional longevity. J. Am. Geriatr. Soc. 59, 1509–1512 (2011).
Bitto, A., Wang, A. M., Bennett, C. F. & Kaeberlein, M. Biochemical genetic pathways that modulate aging in multiple species. Cold Spring Harb. Perspect. Med. 5, a025114 (2015).
Taniguchi, C. M., Emanuelli, B. & Kahn, C. R. Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 (2006).
Kennedy, B. K. & Lamming, D. W. The mechanistic target of rapamycin: the grand conductor of metabolism and aging. Cell Metab. 23, 990–1003 (2016).
Kahn, A. J. FOXO3 and related transcription factors in development, aging, and exceptional longevity. J. Gerontol. A Biol. Sci. Med. Sci. 70, 421–425 (2015).
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
Kops, G. J. et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321 (2002).
Greer, E. L. et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119 (2007).
Cantó, C. et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 (2009).
Brunet, A. et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 (2004).
Yeung, F. et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 (2004).
Tasselli, L., Zheng, W. & Chua, K. F. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28, 168–185 (2017).
Roichman, A. et al. SIRT6 overexpression improves various aspects of mouse healthspan. J. Gerontol. A Biol. Sci. Med. Sci. 72, 603–615 (2017).
Tian, X. et al. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622–638.e622 (2019).
Di Francesco, A., Di Germanio, C., Bernier, M. & de Cabo, R. A time to fast. Science 362, 770–775 (2018).
Mannick, J. B. et al. mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 6, 268ra179 (2014).
Timmers, S. et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 14, 612–622 (2011).
Dai, H., Sinclair, D. A., Ellis, J. L. & Steegborn, C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 188, 140–154 (2018).
van Heemst, D. et al. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell 4, 79–85 (2005).
Milman, S. et al. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 13, 769–771 (2014).
Deelen, J. et al. Gene set analysis of GWAS data for human longevity highlights the relevance of the insulin/IGF-1 signaling and telomere maintenance pathways. Age (Dordr.) 35, 235–249 (2013).
Pawlikowska, L. et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 8, 460–472 (2009).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Hormozdiari, F. et al. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99, 1245–1260 (2016).
Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016).
Holmans, P. et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 85, 13–24 (2009).
Jia, P., Zheng, S., Long, J., Zheng, W. & Zhao, Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 27, 95–102 (2011).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Comput. Biol. 11, e1004219 (2015).
Taşan, M. et al. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat. Methods 12, 154–159 (2015).
Lin, J. R. et al. PGA: post-GWAS analysis for disease gene identification. Bioinformatics 34, 1786–1788 (2018).
Lin, J. R. et al. Integrated Post-GWAS analysis sheds new light on the disease mechanisms of schizophrenia. Genetics 204, 1587–1600 (2016).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Sundaram, L. et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 50, 1161–1170 (2018).
Lin, J. R., Zhang, Q., Cai, Y., Morrow, B. E. & Zhang, Z. D. Integrated rare variant-based risk gene prioritization in disease case-control sequencing studies. PLoS Genet. 13, e1007142 (2017).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Acknowledgements
We thank M. Guo and R. Kohanski at NIA for their scientific input, which contributed to the concepts outlined in this Perspective. This work was supported by grant U19 AG056278 from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Z.D.Z., V.G., W.C.L., L.J.N., Y.S., P.D.R. and J.V. substantially contributed to the discussion and the writing of the content. Z.D.Z., S.M., J.-R.L., S.W., H.Y., N.B., V.G., W.C.L., L.J.N., Y.S., P.D.R. and J.V. researched content for the article, contributed to writing, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
J.V. is a founder of Singulomics Corp. P.R. and L.N. are co-founders of NRTK Biosciences. All other authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Pooja Jha.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Zhang, Z.D., Milman, S., Lin, JR. et al. Genetics of extreme human longevity to guide drug discovery for healthy ageing. Nat Metab 2, 663–672 (2020). https://doi.org/10.1038/s42255-020-0247-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0247-0
This article is cited by
-
Genetik, Epigenetik und Umweltfaktoren der Lebenserwartung – Welche Rolle spielt Nature-versus-Nurture beim Altern?
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2024)
-
Metformin potentiates nephrotoxicity by promoting NETosis in response to renal ferroptosis
Cell Discovery (2023)
-
Induction of proteasomal activity in mammalian cells by lifespan-extending tRNA synthetase inhibitors
GeroScience (2023)
-
High-throughput sequencing analysis of nuclear-encoded mitochondrial genes reveals a genetic signature of human longevity
GeroScience (2023)
-
G protein-coupled receptors that influence lifespan of human and animal models
Biogerontology (2022)