Abstract
The survival and recurrence of dormant tumour cells following therapy is a leading cause of death in patients with cancer. The metabolic properties of these cells are likely to be distinct from those of rapidly growing tumours. Here we show that Her2 downregulation in breast cancer cells promotes changes in cellular metabolism, culminating in oxidative stress and compensatory upregulation of the antioxidant transcription factor NRF2. NRF2 is activated during dormancy and in recurrent tumours in animal models and patients with breast cancer with poor prognosis. Constitutive activation of NRF2 accelerates recurrence, whereas suppression of NRF2 impairs it. In recurrent tumours, NRF2 signalling induces a transcriptional metabolic reprogramming to re-establish redox homeostasis and upregulate de novo nucleotide synthesis. The NRF2-driven metabolic state renders recurrent tumour cells sensitive to glutaminase inhibition, which prevents reactivation of dormant tumour cells in vitro, suggesting that NRF2-high dormant and recurrent tumours may be targeted. These data provide evidence that NRF2-driven metabolic reprogramming promotes the recurrence of dormant breast cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA sequencing data comparing primary and recurrent MTB;TAN-derived tumour cell lines is available online using the National Center for Biotechnology Information (NCBI)Short Read Archive (SRA) under project accession number 626 PRJNA505839 (ref.62). Prognostic analysis for NRF2 gene signatures was performed using Gene expression-based Outcome for Breast cancer Online (GOBO31), which uses four data sets (GSE1456, GSE3494, GSE6532, and GSE7390), available at http://co.bmc.lu.se/gobo/gsa.pl. RNA Sequencing data used for human recurrent breast cancer GSEA were accessed using the NCBI GEO database GSE110590 (ref. 32). Metabolomic data for KEAP1 mutant cancer cell lines was accessed using the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) and supplementary tables from ref. 35. Metabolomics data sets generated in this study are available in Supplementary Table 3.
References
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Boroughs, L. K. & DeBerardinis, R. J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351–359 (2015).
Elstrom, R. L. et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 (2004).
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA 105, 18782–18787 (2008).
Menendez, J. A. et al. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc. Natl Acad. Sci. USA 101, 10715–10720 (2004).
Wang, X., Sun, Y., Wong, J. & Conklin, D. S. PPARγ maintains ERBB2-positive breast cancer stem cells. Oncogene. 32, 5512–5521 (2013).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Tennant, D. A., Duran, R. V., Boulahbel, H. & Gottlieb, E. Metabolic transformation in cancer. Carcinogenesis. 30, 1269–1280 (2009).
Klein, C. A. Framework models of tumor dormancy from patient-derived observations. Curr. Opin. Genet. Dev. 21, 42–49 (2011).
Sosa, M. S., Bragado, P. & Aguirre-Ghiso, J. A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14, 611–622 (2014).
Yeh, A. C. & Ramaswamy, S. Mechanisms of cancer cell dormancy—another hallmark of cancer? Cancer Res. 75, 5014–5022 (2015).
Havas, K. M. et al. Metabolic shifts in residual breast cancer drive tumor recurrence. J. Clin. Invest. 127, 2091–2105 (2017).
Viale, A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014).
Krall, E. B. et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 6, e18970 (2017).
Chio, I. I. C. & Tuveson, D. A. ROS in cancer: the burning question. Trends Mol. Med. 23, 411–429 (2017).
Schafer, Z. T. et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113 (2009).
Moody, S. E. et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, 451–461 (2002).
Moody, S. E. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005).
Alvarez, J. V. et al. Par-4 downregulation promotes breast cancer recurrence by preventing multinucleation following targeted therapy. Cancer Cell 24, 30–44 (2013).
Walens, A et al. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. eLife 8, e43653 (2019).
Feng, Y. et al. SPSB1 promotes breast cancer recurrence by potentiating c-MET signaling. Cancer Discov. 4, 790–803 (2014).
Wang, H. et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430 (2017).
Nguyen, T., Nioi, P. & Pickett, C. B. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 284, 13291–13295 (2009).
DeNicola, G. M. et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011).
Oshimori, N., Oristian, D. & Fuchs, E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015).
Jiang, T. et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 70, 5486–5496 (2010).
Wang, H. et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl Med. 8, 334ra51 (2016).
Kohler, U. A. et al. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology 60, 670–678 (2014).
Kobayashi, M. et al. Identification of the interactive interface and phylogenic conservation of the Nrf2–Keap1 system. Genes Cells 7, 807–820 (2002).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017).
Ringner, M., Fredlund, E., Hakkinen, J., Borg, A. & Staaf, J. GOBO: gene expression-based outcome for breast cancer online. PLoS One 6, e17911 (2011).
Siegel, M. B. et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Invest. 128, 1371–1383 (2018).
Lane, A. N. & Fan, T. W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 43, 2466–2485 (2015).
Mitsuishi, Y. et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012).
Li, H. et al. The landscape of cancer cell line metabolism. Nat. Med. 25, 850–860 (2019).
Sayin, V. I. et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife e28083 (2017).
Furukawa, M. & Xiong, Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25, 162–171 (2005).
Lien, E. C. et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol. 18, 572–578 (2016).
Rada, P. et al. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 31, 1121–1133 (2011).
Piskounova, E. et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191 (2015).
Harris, I. S. et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 27, 211–222 (2015).
Deblois, G. et al. ERRα mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat. Commun. 7, 12156 (2016).
Takahashi, N. et al. Cancer cells co-opt the neuronal redox-sensing channel TRPA1 to promote oxidative-stress tolerance. Cancer Cell. 33, 985–1003.e7 (2018).
Park, S. et al. ERRα-regulated lactate metabolism contributes to resistance to targeted therapies in breast cancer. Cell Rep. 15, 323–335 (2016).
DeNicola, G. M. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015).
Padmanabhan, B. et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700 (2006).
Hast, B. E. et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 74, 808–817 (2014).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Nagini, S., Sophia, J. & Mishra, R. Glycogen synthase kinases: moonlighting proteins with theranostic potential in cancer. Semin. Cancer Biol. 56, 25–36 (2019).
Muir, A., Danai, L. V. & Vander Heiden, M. G. Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis. Model. Mech. 11, dmm035758 (2018).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 (2009).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science 307, 1098–1101 (2005).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Liu, X., Ser, Z. & Locasale, J. W. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem. 86, 2175–2184 (2014).
Liberti, M. V. et al. A predictive model for selective targeting of the warburg effect through GAPDH inhibition with a natural product. Cell Metab. 26, 648–659.e8 (2017).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Meier, J. A., Zhang, F. & Sanjana, N. E. GUIDES: sgRNA design for loss-of-function screens. Nat. Methods 14, 831–832 (2017).
Chen, S. et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Mabe, N. W. et al. G9a promotes breast cancer recurrence through repression of a pro-inflammatory program. Preprint at bioRxiv, https://doi.org/10.1101/2020.01.09.900183 (2020).
Acknowledgements
We thank S. Y. Kim from the Duke Functional Genomics Core, N. Devos from the Duke Center for Genomic and Computational Biology and Y. Gao from the Duke Light Microscopy Core. We would also like to thank N. Mabe for his advice on bioinformatic analysis. This work was funded by National Cancer Institute grants R01CA208042 (J.V.A.), R01CA193256 (J.W.L.), F31CA228321 (D.B.F.), the V-Foundation, Golfers Against Cancer, the Integrative metabolomics shared resource, and by startup funds from the Duke Cancer Institute, the Duke University School of Medicine and the Whitehead Foundation (to J.V.A.).
Author information
Authors and Affiliations
Contributions
J.V.A. and D.B.F. were responsible for the conception, design and interpretation of all experiments. M.D.H assisted in the conception and interpretation of metabolic experiments. D.B.F., N.M.G.G., B.J.M., R.L., L.C.N. and R.N. performed experiments and collected data. J.L. and J.W.L. designed, performed, and analysed the metabolomics experiments. D.B.F. and J.V.A. wrote the manuscript. J.V.A. supervised all work.
Corresponding author
Ethics declarations
Competing interests
J.W.L. advises Nanocare Technologies, Raphael Pharmaceuticals, and Restoration Foodworks. All other authors report no conflicts.
Additional information
Peer review information Primary handling editors: Ana Mateus; Elena Bellafante.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
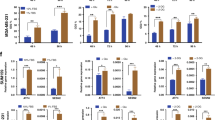
Extended Data Fig. 1 Signalling changes induced by Her2 downregulation.
a, Western blots for p-Her2 (Tyr1221/1222), p-ERK1/2 (Thr202/Tyr204), p-Akt (Ser473), and p-S6-RP (Ser240/244) in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off). Data represent 3 independent experiments. b, Western blots for Her2, ERK1/2, Akt, and PCNA in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off). Data represent 3 independent experiments. c, Western blot for phosphorylated pyruvate dehydrogenase (p-PDH) (Ser293) in mammospheres cultured in the presence of dox (Her2 on) or without dox (Her2 off) for 4 and 14 days. Data represent 3 independent experiments. d, Relative ratio of oxidised (NADP+) to reduced NADP (NADPH) between Her2 off and Her2 on conditions. For –dox cells, the NADP+ :NADPH ratio is shown for a single sample, since the NADPH level for the other samples was below the limit of detection, and therefore the ratio is undefined.
Extended Data Fig. 2 Her2-targeted therapies induce ROS in breast cancer cells.
a, DCFDA staining showing ROS levels in mammospheres cultured with dox or without dox for 2 days. Significance was determined by two-sided Student’s t test, and data are represented as mean ± SEM (n = 6 fields of view). Scale bars represent 100μm. b, DCFDA staining showing ROS levels in MDA-MB-231 cells treated with 500 nM lapatinib for 48 hours. Data are representative of n = 3 biologically independent replicates for a single experiment. c, DCFDA staining showing ROS levels in SKBR3, BT474, and MDA-MB-231 cells treated with 500 nM neratinib for 48 hours. Data are shown as the mean ± SEM for n = 3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
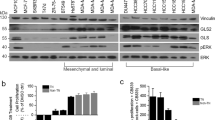
Extended Data Fig. 3 PI3K inhibition induces ROS in breast cancer cells.
Relative viability of HCC1954 cells treated with lapatinib and 5 mM NAC for 72 hours. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). b, DCFDA staining showing ROS levels in BT474 and SKBR3 cells treated with lapatinib or 100 μM dehydroepiandrosterone (DHEA) for 48 hours. BT474 cells were treated with 100 μM tert-butyl hydrogen peroxide (TBHP) for 12 hours as a positive control. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 biologically independent experiments. c, DCFDA staining showing ROS levels in BT474 cells treated with lapatinib and 50 μM 6-aminonicotinamide (6-An) for 48 hours. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 biologically independent experiments. d, DCFDA staining showing ROS levels in mammospheres treated with 50 μM 6-An or 100 μM TBHP for 24 hours. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 biologically independent experiments. e, Relative viability of mammospheres cultured with or without dox in the presence or absence of 100 μM etomoxir (eto) for 7 days. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons test. f, qRT-PCR analysis of Cpt1a, Cpt1b, and CD36 expression in 2 independent mammosphere cultures grown in the presence of dox or without dox for 4 days. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA with Tukey’s multiple comparisons test. g, Boron-dipyrromethene (BODIPY) staining showing lipid droplets (green) in mammospheres cultured in the presence of dox. Scale bar, 25 μm. h, DCFDA staining showing ROS levels in T47D, BT474, MDA-MB-231, and SKBR3 cells treated with 500 nM BKM120 for 48 hours. Data are representative of n = 3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test. i, DCFDA staining showing ROS levels in MDA-MB-231 cells treated with 500 nM JQ1 for 48 hours. Data are representative of n = 3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 4 DNA damage therapy induces ROS in breast cancer cells.
a, DCFDA staining showing ROS levels in T47D, BT474, and MCF7 cells treated with 1 μM fulvestrant for 48 hours. Data are representative of n = 3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test. b, DCFDA staining showing ROS levels in MCF7, SKBR3, MDA-MB-231, BT474, and T47D cells treated with indicated doses of etoposide for 48 hours. Data are representative of n = 3 biologically independent replicates for a single experiment. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 5 Dormant and recurrent tumour cells upregulate NRF2.
a, qRT-PCR analysis of Gclm, Nqo1, and Hmox1 expression in primary tumours (n = 5) and tumours 48 hours after dox withdrawal (Her2 off; n = 4). Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). b, Hematoxylin and eosin staining of a mammary gland with residual tumours, indicated by black arrowheads. LN indicates the lymph node. Black box on left panel indicates magnified residual tumour shown in right panel. Scale bars represent 2 mm (left) and 200μm (right). Data are representative of 3 biologically independent experiments. c, Hematoxylin and eosin staining (top panels) and Ki67 immunohistochemical staining (brown, middle and bottom panels) of serial tissue sections from a representative primary tumour and representative residual tumours. White boxes indicate magnified section shown in bottom panels. Scale bars represent 200μm (top and middle panels) and 50μm (bottom panels). d, Quantification of the proportion of Ki67-positive cells identified by immunohistochemical staining of primary (n = 4) and residual (n = 6) tumours. Significance was determined by two-sided Student’s t test. e, Waterfall plot of RNA sequencing data showing the log2(fold change) in expression of NRF2 target genes between primary tumour cell lines (n = 2) and recurrent tumour cell lines (n = 2). Red bars indicate genes whose expression is greater than 1.5-fold higher in recurrent cells.
Extended Data Fig. 6 NRF2 expression and activity during tumour recurrence.
a, qRT-PCR analysis of Nqo1, Slc7a11, Gclm, and Hmox1 expression in primary tumour cells expressing an empty vector (Control) or caNRF2. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). Data are representative of 2 independent experiments. b, DCFDA staining showing ROS levels in primary tumour cells expressing an empty vector (Cont) or caNRF2. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 independent experiments. c, Kaplan-Meier plot showing primary tumour-free survival for mice bearing control tumours (n = 27 independent tumours) or caNRF2 tumours (n = 32 independent tumours). Hazard ratio and P value were determined by two-sided log-rank (Mantel-Cox) test. d, Western blot for NRF2 and NQO1 in caNRF2-expressing primary and recurrent tumours. Blot is representative of a single experiment. e, Western blots for NRF2 and NQO1 in control primary and recurrent tumours. Blot is representative of a single experiment.
Extended Data Fig. 7 NRF2 is upregulated in human recurrent breast tumours.
a, Gene set enrichment analysis of matched primary and recurrent breast tumours (n = 15 patients) showing enrichment of ‘Reactive Oxygen Species Pathway’ signature in recurrent tumours. b, Gene set enrichment analysis of matched primary and recurrent breast tumours (n = 15 patients) showing enrichment of “Fatty Acid Oxidation” signature in recurrent tumours. c, Gene set enrichment analysis of matched primary and recurrent breast tumours (n = 15 tumours) showing non-significant of the “Notch Signaling” signature in recurrent tumours. d, Volcano plot showing enrichment scores for the “Notch Signaling” signature in recurrent tumours in n = 15 individual patients. e, The percentage of patients whose recurrent tumours had a significantly enriched (red), unchanged (grey), or significantly decreased (blue) “Notch Signaling” signature. f, Gene set enrichment analysis of matched primary and recurrent breast tumours (n = 15 tumours) showing non-significant of the “Wnt Beta Catenin Signaling” signature in recurrent tumours. g, Volcano plot showing enrichment scores for the “Wnt Beta Catenin Signaling” signature in recurrent tumours in n = 15 individual patients. h, The percentage of patients whose recurrent tumours had a significantly enriched (red), unchanged (grey), or significantly decreased (blue) “Wnt Beta Catenin Signaling” signature. i, Expression of NRF2, KEAP1, TBP, and 10 canonical NRF2 target genes in matched primary and recurrent tumours from representative patients. j, qRT-PCR analysis showing expression of NRF2 (Nfe2l2) and its target genes, Slc7a11, Nqo1, Hmox1, Gclm, Txnrd1, and Pgd, in shNRF2 #1 and shNRF2 #2 injected cells and tumour samples from Fig. 5d, e relative to shScramble control. a–c, f, Enrichment scores were calculated using the Kolmogorov-Smirnov statistic and P values were calculated using permutation testing with 1000 permutations.
Extended Data Fig. 8 NRF2 has limited effects on metabolism in primary tumour cells.
a, Relative GSH:GSSG ratio in primary tumour cells expressing empty vector (control) or caNRF2. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two sided Student’s t test. b–d, qRT-PCR analysis of Gclm and Slc7a11 (b), Txnrd1 (c), and Pgd (d) expression in control (shScr) and NRF2-knockdown (shNRF2) primary tumour cell lines. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test). e, Relative levels of the PPP metabolites 6-phosphogluconate (6PG) and ribose 5-phosphate (R5P) in primary tumour cells expressing an empty vector (control) or caNRF2. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. p values were determined by two-sided Student’s t test. f, Waterfall plot showing the fold change in nucleotide levels between primary tumour cells expressing an empty vector (control) or caNRF2. g, Relative levels of the PPP metabolites 6PG and R5P in primary tumour cells cultured with dox (Her2 on) and without dox for 7 days (Her2 off). Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-sided Student’s t test.
Extended Data Fig. 9 Identification of essential NRF2-regulated pathways in recurrent tumours.
a, Table showing the fold change (FC, injected cells/tumours) and NRF2-regulated pathway for all genes tested for both cell lines (n = 5 tumours for recurrent tumour line 1; n = 12 for recurrent tumour line 2). Significance was determined using Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK). b, Average reads per gene for injected cell samples for each recurrent cell line. Genes in NRF2 regulated pathways are represented as hollow circles. Essential ribosomal genes are represented as red dots. Non-targeting sgRNA are represented as blue dots. Genes are ranked from fewest to most reads. c, Dose-response curves showing relative viability of two primary and two recurrent tumour cell lines treated with indicated doses of BPTES for 48 hours. Data points represent mean ± SEM for n = 3 biologically independent replicates. d, Dose-response curves showing relative viability of control (shScr) and NRF2-knockdown (shNRF2) recurrent tumour cells treated with indicated doses of BPTES for 72 hours. Data points represent mean ± SEM for n = 3 biologically independent replicates. e, Dose response curves showing relative viability of primary tumour cells expressing an empty vector (control) or constitutively active NRF2 (caNRF2) treated with indicated doses of BPTES for 72 hours. Data points represent mean ± SEM for n = 3 biologically independent replicates. f, DCFDA staining showing ROS levels in control (shScr) and NRF2-knockdown (shNRF2) recurrent cells treated with 3 μM BPTES for 48 hours. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 independent experiments. g, Relative viability of recurrent cell lines treated with CB-839 and dimethyl-α-ketoglutarate (dm-αKG), as indicated. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. Significance was determined by two-way ANOVA (Tukey’s multiple comparisons test).
Extended Data Fig. 10 Mechanism of NRF2 activation in recurrent tumours.
a, qRT-PCR analysis of NRF2 (Nfe2l2;) expression in two independent primary cell lines and two independent recurrent (Rec) cell lines. Data are shown as the mean ± SEM for n = 3 biologically independent replicates. b, qRT-PCR showing mRNA expression of Keap1 in two independent primary and recurrent (Rec) cell lines. Data are shown as the mean of n = 2 biologically independent replicates and are representative of 2 independent experiments. c, Western blot for KEAP1 in two independent primary cell lines and two independent recurrent (Rec) cell lines. Blot is representative of 3 independent experiments. d, Western blots for NRF2 and NQO1 in a primary cell line and a recurrent cell line treated with the indicated dose of TBHQ for 16 hours. Blot is representative of 2 independent experiments. e, Quantification of NRF2 expression (normalised to Tubulin) from (d). f, Western blot showing p-Akt (Ser473) levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 30 minutes. Blot is representative of 2 independent experiments. g, Western blot showing p-Akt (Ser473), p-GSK-3β (Ser9), and p-PRAS40 levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 3 days. Blot is representative of 2 independent experiments. h, Western blot showing NRF2 and NQO1 protein levels in two recurrent cell lines treated with the Akt inhibitor, MK2206 (MK), for 3 days. Blot is representative of 2 independent experiments. i, Western blot showing p-GSK-3β (S9) levels in two independent primary cell lines and two independent recurrent cell lines. Blot is representative of 2 independent experiments.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Supplementary Table 3
Unprocessed metabolomics data
Source data
Source Data Fig. 2
FACS gating strategy
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 4
Unprocessed Western Blots
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 8
Unprocessed Western Blots
Source Data Extended Data Fig. 1
Unprocessed Western Blots
Source Data Extended Data Fig. 5
Unprocessed Western Blots
Source Data Extended Data Fig. 10
Unprocessed Western Blots
Rights and permissions
About this article
Cite this article
Fox, D.B., Garcia, N.M.G., McKinney, B.J. et al. NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nat Metab 2, 318–334 (2020). https://doi.org/10.1038/s42255-020-0191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0191-z
This article is cited by
-
Metabolic adaptation towards glycolysis supports resistance to neoadjuvant chemotherapy in early triple negative breast cancers
Breast Cancer Research (2024)
-
The pleiotropic functions of reactive oxygen species in cancer
Nature Cancer (2024)
-
How much do we know about the metastatic process?
Clinical & Experimental Metastasis (2024)
-
TH-4000, a hypoxia-activated pan-HER inhibitor, shows excellent preclinical efficacy for the treatment of HER2+ breast cancer
Archives of Toxicology (2024)
-
Investigation of the effects of catharanthine and Q10 on Nrf2 and its association with MMP-9, MRP1, and Bcl-2 and apoptosis in a model of hepatocellular carcinoma
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)