Abstract
The neonatal mammalian heart is capable of regeneration for a brief window of time after birth. However, this regenerative capacity is lost within the first week of life, which coincides with a postnatal shift from anaerobic glycolysis to mitochondrial oxidative phosphorylation, particularly towards fatty-acid utilization. Despite the energy advantage of fatty-acid beta-oxidation, cardiac mitochondria produce elevated rates of reactive oxygen species when utilizing fatty acids, which is thought to play a role in cardiomyocyte cell-cycle arrest through induction of DNA damage and activation of DNA-damage response (DDR) pathway. Here we show that inhibiting fatty-acid utilization promotes cardiomyocyte proliferation in the postnatal heart. First, neonatal mice fed fatty-acid-deficient milk showed prolongation of the postnatal cardiomyocyte proliferative window; however, cell-cycle arrest eventually ensued. Next, we generated a tamoxifen-inducible cardiomyocyte-specific pyruvate dehydrogenase kinase 4 (PDK4) knockout mouse model to selectively enhance oxidation of glycolytically derived pyruvate in cardiomyocytes. Conditional PDK4 deletion resulted in an increase in pyruvate dehydrogenase activity and consequently an increase in glucose relative to fatty-acid oxidation. Loss of PDK4 also resulted in decreased cardiomyocyte size, decreased DNA damage and expression of DDR markers and an increase in cardiomyocyte proliferation. Following myocardial infarction, inducible deletion of PDK4 improved left ventricular function and decreased remodelling. Collectively, inhibition of fatty-acid utilization in cardiomyocytes promotes proliferation, and may be a viable target for cardiac regenerative therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-seq raw data have been deposited in the NCBI Sequence Read Archive (SRA) database with accession number PRJNA593900. Source Data for Figs. 3 and 4 and Extended Data Fig. 4 are available online.
References
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009).
Mahmoud, A. I. et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249–253 (2013).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Becker, R. O., Chapin, S. & Sherry, R. Regeneration of the ventricular myocardium in amphibians. Nature 248, 145–147 (1974).
Oberpriller, J. O. & Oberpriller, J. C. Response of the adult newt ventricle to injury. J. Exp. Zool. 187, 249–253 (1974).
Neff, A. W., Dent, A. E. & Armstrong, J. B. Heart development and regeneration in urodeles. Int. J. Dev. Biol. 40, 719–725 (1996).
Flink, I. L. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat. Embroyl. (Berl). 205, 235–244 (2002).
Jopling, C. et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 (2010).
Kikuchi, K. et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605 (2010).
Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M. & Field, L. J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 271, H2183–H2189 (1996).
Nassar, R., Reedy, M. C. & Anderson, P. A. Developmental changes in the ultrastructure and sarcomere shortening of the isolated rabbit ventricular myocyte. Circ. Res. 61, 465–483 (1987).
Wu, Y. & Wu, E. X. MR study of postnatal development of myocardial structure and left ventricular function. J. Magn. Reson. Imaging 30, 47–53 (2009).
Sdek, P. et al. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J. Cell. Biol. 194, 407–423 (2011).
Xin, M. et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl Acad. Sci. 110, 13839–13844 (2013).
Chen, J. et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566 (2013).
Eulalio, A. et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381 (2012).
Bersell, K., Arab, S., Haring, B. & Kuhn, B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270 (2009).
Porrello, E. R. et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 109, 670–679 (2011).
Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science 298, 2188–2190 (2002).
Bergmann, O. et al. Cardiomyocyte renewal in humans. Circ. Res. 110, e17–e18 (2012).
Laflamme, M. A., Myerson, D., Saffitz, J. E. & Murry, C. E. Evidence for cardiomyocyte repopulation by extracardiac progenitors in transplanted human hearts. Circ. Res. 90, 634–640 (2002).
Nadal-Ginard, B. [Generation of new cardiomyocytes in the adult heart: Prospects of myocardial regeneration as an alternative to cardiac transplantation]. Rev. Esp. Cardiol. 54, 543–550 (2001).
Quaini, F. et al. Chimerism of the transplanted heart. N. Engl. J. Med. 346, 5–15 (2002).
Lam, N. T. & Sadek, H. A. Neonatal heart regeneration. Circulation 138, 412–423 (2018).
Elhelaly, W. M. et al. C-Kit cells do not significantly contribute to cardiomyogenesis during neonatal heart regeneration. Circulation 139, 559–561 (2019).
Porrello, E. R. et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl Acad. Sci. USA 110, 187–192 (2013).
Zhu, W. et al. Regenerative potential of neonatal porcine hearts. Circulation 138, 2809–2816 (2018).
Ye, L. et al. Early regenerative capacity in the porcine heart. Circulation 138, 2798–2808 (2018).
Puente, B. N. et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 565–579 (2014).
Fisher, D. J., Heymann, M. A. & Rudolph, A. M. Myocardial oxygen and carbohydrate consumption in fetal lambs in utero and in adult sheep. Am. J. Physiol. 238, H399–H405 (1980).
Lopaschuk, G. D., Collins-Nakai, R. L. & Itoi, T. Developmental changes in energy substrate use by the heart. Cardiovasc. Res. 26, 1172–1180 (1992).
Wisneski, J. A. et al. Metabolic fate of extracted glucose in normal human myocardium. J. Clin. Invest. 76, 1819–1827 (1985).
Gertz, E. W., Wisneski, J. A., Stanley, W. C. & Neese, R. A. Myocardial substrate utilization during exercise in humans. Dual carbon-labeled carbohydrate isotope experiments. J. Clin. Invest. 82, 2017–2025 (1988).
Lehman, J. J. & Kelly, D. P. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin. Exp. Pharamcol. Physiol. 29, 339–345 (2002).
Anderson, S. M., Rudolph, M. C., McManaman, J. L. & Neville, M. C. Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 9, 204 (2007).
Rindler, P. M., Crewe, C. L., Fernandes, J., Kinter, M. & Szweda, L. I. Redox regulation of insulin sensitivity due to enhanced fatty acid utilization in the mitochondria. Am. J. Physiol. Heart Circ. Physiol. 305, H634–H643 (2013).
Bowker-Kinley, M. M., Davis, W. I., Wu, P., Harris, R. A. & Popov, K. M. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 329 (Pt. 1), 191–196 (1998).
Rardin, M. J., Wiley, S. E., Naviaux, R. K., Murphy, A. N. & Dixon, J. E. Monitoring phosphorylation of the pyruvate dehydrogenase complex. Anal. Biochem. 389, 157–164 (2009).
Sugden, M. C. & Holness, M. J. Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Arch. Physiol. Biochem. 112, 139–149 (2006).
Holness, M. J., Kraus, A., Harris, R. A. & Sugden, M. C. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes 49, 775–781 (2000).
Holness, M. J. et al. Evaluation of the role of peroxisome-proliferator-activated receptor alpha in the regulation of cardiac pyruvate dehydrogenase kinase 4 protein expression in response to starvation, high-fat feeding and hyperthyroidism. Biochem. J. 364, 687–694 (2002).
Anderson, E. J., Yamazaki, H. & Neufer, P. D. Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. J. Biol. Chem. 282, 31257–31266 (2007).
Rindler, P. M., Plafker, S. M., Szweda, L. I. & Kinter, M. High dietary fat selectively increases catalase expression within cardiac mitochondria. J. Biol. Chem. 288, 1979–1990 (2013).
Seifert, E. L., Estey, C., Xuan, J. Y. & Harper, M. E. Electron transport chain-dependent and -independent mechanisms of mitochondrial H2O2 emission during long-chain fatty acid oxidation. J. Biol. Chem. 285, 5748–5758 (2010).
Bobrovnikova-Marjon, E. et al. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc. Natl Acad. Sci. USA 105, 16314–16319 (2008).
Wagner, K.-U., Ward, T., Davis, B., Wiseman, R. & Hennighausen, L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 10, 545–553 (2001).
Ali, S. R. et al. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc. Natl Acad. Sci. USA 111, 8850–8855 (2014).
Smagris, E. et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61, 108–118 (2015).
Lewandowski, E. D. & White, L. T. Pyruvate dehydrogenase influences postischemic heart function. Circulation 91, 2071–2079 (1995).
Ussher, J. R. et al. Stimulation of glucose oxidation protects against acute myocardial infarction and reperfusion injury. Cardiovasc. Res. 94, 359–369 (2012).
Dyck, J. R. et al. Absence of malonyl coenzyme A decarboxylase in mice increases cardiac glucose oxidation and protects the heart from ischemic injury. Circulation 114, 1721–1728 (2006).
Liu, Z. et al. PDK4 protein promotes tumorigenesis through activation of cAMP-response element-binding protein (CREB)-Ras homolog enriched in brain (RHEB)-mTORC1 signaling cascade. J. Biol. Chem. 289, 29739–29749 (2014).
Thoudam, T. et al. PDK4 augments ER–mitochondria contact to dampen skeletal muscle insulin signaling during obesity. Diabetes 68, 571–586 (2018).
Sohal, D. S. et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89, 20–25 (2001).
Harding, H. P., Zhang, Y. & Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 (1999).
Cavener, D. R., Gupta, S. & McGrath, B. C. PERK in beta cell biology and insulin biogenesis. Trends. Endocrinol. Metab. 21, 714–721 (2010).
Zhang, P. et al. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22, 3864–3874 (2002).
Matsuhashi, T. et al. Activation of pyruvate dehydrogenase by dichloroacetate has the potential to induce epigenetic remodeling in the heart. J. Mol. Cell. Cardiol. 82, 116–124 (2015).
Friedewald, W. T., Levy, R. I. & Fredrickson, D. S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18, 499–502 (1972).
Ejsing, C. S. et al. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 78, 6202–6214 (2006).
Acknowledgements
H. I. May (Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, Texas, USA) for the myocardial infarction surgeries in mice. H.A.S. is supported by grants from the NIH (1R01HL115275 and 5R01H2131778), National Aeronautics and Space Administration (NNX-15AE06G), American Heart Association (16EIA27740034), Cancer Prevention and Research Institute of Texas (RP160520), Hamon Center for Regenerative Science and Medicine and Fondation Leducq. N.T.L. is supported by a Haberecht Wildhare-Idea Research Grant. A.D.S. is supported by grant from the NIH (R37-HL034557), C.R.M. is supported by grant from the NIH (P41-EB015908) and G.S. is supported by the grant from the AHA (18POST34050049). M.K. is supported by the grants from NIH (3P20GM103447 and 5P30AG050911). I.M.M. is supported by Alfonso Martin Escudero Foundation Fellowship. N.U.N.N. is supported by AHA Postdoctoral Fellowship 19POST34450039. J.A.H. is supported by grants from the NIH (HL-120732, HL-128215 and HL-126012).
Author information
Authors and Affiliations
Contributions
A.C.C. and N.T.L. performed the experiments. A.C.C, N.T.L. and H.A.S. designed the experiments, performed the analyses and wrote the manuscript. J.J.S. contributed to writing and manuscript preparation. J.J.S. and S.L. performed the echocardiography. A.E. and L.I.S. performed PDH activity analysis. G.V., K.M.E., M.A.M. and J.G.M. performed the lipidomics experiments. Y.N., S.A. and A.A. performed the DNA-damage experiments. I.M.M., L.H. and P.W.S performed the lipid serum profile experiments. G.S., A.D.S., C.R.M. and C.K. performed the NMR experiments. M.T.K. performed the proteomics data. W.L.W.T., C.G.A.N. and R.S.Y.F. performed the RNA-seq. C.X. and M.K. contributed to the bioinformatics analysis. A.H.M.P., N.U.N.N., M.S.A. and W.M.E contributed and performed the experiments in mice. A.H.M.P., E.L.E and U.B.P. contributed to the experimental design and performed the experiments in mice. J.A.H. contributed to experimental design in mice. L.I.S. and H.A.S. conceived the project and contributed to manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quantitative mass spectrometry analysis of fatty acids biosynthesis liver enzymes.
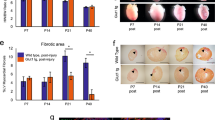
a, Schematic view of cholesterol and triglyceride biosynthesis pathways. Red, green and gray colors represent the upregulated, downregulated and unchanged protein expression, respectively. The analysis shows a significant increase in the enzymes involved in the synthesis of saturated fatty acids and triglycerides in FDM compare to CM mice. Enzymes involved in cholesterol biosynthesis were downregulated in the FDM liver. b, Table showing the average, fold change and p-value of the hepatic enzymes involved in fatty acid biosynthesis, identified by quantitative mass spectrometry between FDM and CM mice (n=4 biologically independent mice per group). CM, control milk group; FDM, fat deficient milk group. Statistical analysis was performed with two-tailed Student’s t-test.
Extended Data Fig. 2 Serum lipids and blood glucose measurements.
a, At 12 days postnatally, beyond lactation, pups were exposed to a regular diet or Fat Free Diet. Samples were collected at 10 weeks postnatally. Data in b-f represent the blood glucose, total cholesterol, triglycerides, HDL cholesterol or LDL cholesterol measurements, respectively. (n=7 biologically independent mice per group) Data presented as the mean ± s.e.m. Statistical analysis was analyzed with two-tailed Student’s t-test.
Extended Data Fig. 3 RNA-seq analysis of PDK4 KO and Control hearts.
a, Heat-map of all dysregulated genes. Yellow and blue colors represent upregulated and downregulated genes, respectively (n=5 biologically independent mice for control and n=4 biologically independent mice for PDK4 KO group). b, Scatter plot showing the Fragments Per Kilobase of transcript, per Million mapped reads (FPKM) in control and PDK4 KO group (n=5 biologically independent mice for control and n=4 biologically independent mice for PDK4 KO). c, Volcano plot of the log of fold change dysregulated genes between Control and PDK4 KO group. (n=5 biologically independent mice for control and n=4 biologically independent mice for PDK4 KO). Statistic analysis was performed by two-tailed F-test for differential gene expression analysis. d, Ontology analysis performed using DAVID Functional Annotation Tool. Statistics analysis was performed by hypergeometric test. e, heat maps showing a number of dysregulated pathways, including DNA replication, lipid metabolic process, carbohydrate metabolic process, cell cycle and cell growth. Red and green colors represent upregulated and downregulated genes, respectively (n=5 biologically independent mice for control and n=4 biologically independent mice for PDK4 KO). CT: Control.
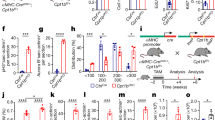
Extended Data Fig. 4 Timeline of PDK4 deletion in the inducible model.
Western blot of PDK4 shows elimination of the protein as early as 4 days following the first tamoxifen injection.
Extended Data Fig. 5 Echocardiography parameters.
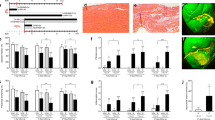
a, left ventricular internal dimension in diastole (LVIDd); b, left ventricular internal dimension in systole (LVIDs), echocardiogram measurements, comparing PDK4 KO and the control αMHC-MerCreMer (MCM) at different time points (n=5 biologically independent mice per group). Data are represented as the mean ± s.d. Analyses were performed by unpaired two-tailed Student’s t-test to compare MCM vs PDK4 KO. Paired two-tailed Student’s t-test was used to compare two time points within the same mouse (6 months post MI versus 1 week post MI). c, LVDd; d, LVIDs echocardiogram measurements, comparing uninjured versus myocardial infarction (MI) MCM and PDK4 KO groups (n=4 biologically independent mice for uninjured and n=5 biologically independent mice for MI groups). Data are represented as the mean ± s.e.m. Analyses were performed by Two-way ANOVA followed by Tukey’s multiple comparisons test, with individual variance computed for each comparison. ns, not significant. e, Trichrome staining of control MCM and PDK4 KO uninjured hearts, 6 months post Tamoxifen injection. Images are representative of three independently performed experiments, with similar results.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Source data
Source Data Fig. 3
Unprocessed western blot referent to figure 3
Source Data Fig. 4
Unprocessed western blot referent to figure 4
Source Data Fig. Extended Data Fig. 4
Unprocessed western blot referent to extended data fig. 4
Rights and permissions
About this article
Cite this article
Cardoso, A.C., Lam, N.T., Savla, J.J. et al. Mitochondrial substrate utilization regulates cardiomyocyte cell-cycle progression. Nat Metab 2, 167–178 (2020). https://doi.org/10.1038/s42255-020-0169-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0169-x
This article is cited by
-
Mitochondrial-to-nuclear communications through multiple routes regulate cardiomyocyte proliferation
Cell Regeneration (2024)
-
Cas13b-mediated RNA targeted therapy alleviates genetic dilated cardiomyopathy in mice
Cell & Bioscience (2024)
-
YAP induces a neonatal-like pro-renewal niche in the adult heart
Nature Cardiovascular Research (2024)
-
Animal models to study cardiac regeneration
Nature Reviews Cardiology (2024)
-
Molecular mechanisms of ferroptosis in cardiovascular disease
Molecular and Cellular Biochemistry (2024)