Abstract
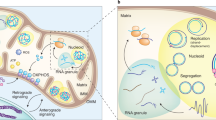
The mammalian genome comprises nuclear DNA (nDNA) derived from both parents and mitochondrial DNA (mtDNA) that is maternally inherited and encodes essential proteins required for oxidative phosphorylation. Thousands of copies of the circular mtDNA are present in most cell types that are packaged by TFAM into higher-order structures called nucleoids1. Mitochondria are also platforms for antiviral signalling2 and, due to their bacterial origin, mtDNA and other mitochondrial components trigger innate immune responses and inflammatory pathology2,3. We showed previously that cytoplasmic release of mtDNA activates the cGAS–STING–TBK1 pathway resulting in interferon-stimulated gene (ISG) expression that promotes antiviral immunity4. Here, we find that persistent mtDNA stress is not associated with basally activated NF-κB signalling or interferon gene expression typical of an acute antiviral response. Instead, a specific subset of ISGs that includes Parp9 remains activated by the unphosphorylated form of ISGF3 that enhances nDNA damage and repair responses. In cultured primary fibroblasts and cancer cells, the chemotherapeutic drug doxorubicin causes mtDNA damage and release, which leads to cGAS–STING–dependent ISG activation. In addition, mtDNA stress in TFAM-deficient mouse melanoma cells produces tumours that are more resistant to doxorubicin in vivo. Finally, Tfam+/− mice exposed to ionizing radiation exhibit enhanced nDNA repair responses in spleen. Therefore, we propose that damage to and subsequent release of mtDNA elicits a protective signalling response that enhances nDNA repair in cells and tissues, suggesting that mtDNA is a genotoxic stress sentinel.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
CCLE RNA-seq data can be found through the link https://portals.broadinstitute.org/ccle. TFAM microarray data are available in the NCBI GEO database with accession number GSE63767. Source data for Figs. 1–4 and Extended Data Figs. 1–9 are provided with the paper. Additional qPCR biological replicate data can be found in the supplementary files. Other data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Code availability
Code used for data analysis was deposited in GitHub: http://github.com/kepbod/CCLE_analysis
References
Bogenhagen, D. F., Rousseau, D. & Burke, S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675 (2008).
West, A. P., Shadel, G. S. & Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 11, 389–402 (2011).
West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17, 363–375 (2017).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Zhong, Z. et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 560, 198–203 (2018).
Aarreberg, L. D. et al. Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol. Cell 74, 801–815 (2019). e806.
Bai, J. et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl Acad. Sci. USA 114, 12196–12201 (2017).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014).
Grandvaux, N. et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76, 5532–5539 (2002).
Honda, K. & Taniguchi, T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658 (2006).
Cheon, H. et al. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32, 2751–2763 (2013).
Cheon, H. & Stark, G. R. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl Acad. Sci. USA 106, 9373–9378 (2009).
Sung, P. S. et al. Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc. Natl Acad. Sci. USA 112, 10443–10448 (2015).
Weichselbaum, R. R. et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl Acad. Sci. USA 105, 18490–18495 (2008).
Minn, A. J. Interferons and the immunogenic effects of cancer therapy. Trends Immunol. 36, 725–737 (2015).
Yang, C. S. et al. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516 (2017). e505.
Yan, Q. et al. BAL1 and its partner E3 ligase, BBAP, link poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8. Mol. Cell. Biol. 33, 845–857 (2013).
Yan, Q. et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell 36, 110–120 (2009).
Zhang, Y. et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 16, 1215–1227 (2015).
Abe, T. & Barber, G. N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 (2014).
Buondonno, I. et al. Mitochondria-targeted doxorubicin: a new therapeutic strategy against doxorubicin-resistant osteosarcoma. Mol. Cancer Ther. 15, 2640–2652 (2016).
Wang, J. et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30, 428–435 (2017).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Singh, G., Sharkey, S. M. & Moorehead, R. Mitochondrial DNA damage by anticancer agents. Pharmacol. Ther. 54, 217–230 (1992).
Hämäläinen, R. H. et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nat. Metab. 1, 958–965 (2019).
Quiros, P. M., Mottis, A. & Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226 (2016).
Scheibye-Knudsen, M., Fang, E. F., Croteau, D. L., Wilson, D. M. III & Bohr, V. A. Protecting the mitochondrial powerhouse. Trends Cell Biol. 25, 158–170 (2015).
Shadel, G. S. & Clayton, D. A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66, 409–435 (1997).
Woo, D. K. et al. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC Min/+ mice. Am. J. Pathol. 180, 24–31 (2012).
Durbin, J. E., Hackenmiller, R., Simon, M. C. & Levy, D. E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Yousefzadeh, M. J. et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014).
Oeck, S. et al. The Focinator v2-0 – graphical interface, four channels, colocalization analysis and cell phase identification. Radiat. Res. 188, 114–120 (2017).
Oeck, S., Malewicz, N. M., Hurst, S., Rudner, J. & Jendrossek, V. The Focinator – a new open-source tool for high-throughput foci evaluation of DNA damage. Radiat. Oncol. 10, 163 (2015).
Furda, A., Santos, J. H., Meyer, J. N. & Van Houten, B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 1105, 419–437 (2014).
Acknowledgements
We thank A. Iwasaki and C. Dela-Cruz for reagents and advice, A. Mennone Jr. and T. Zhang for assistance with microscopy, C. O’Connor for help with cell sorting and flow cytometry, M. Leblanc and Y.-L. Chang for mouse tissue collection, S. O. Kelley and T. Sack for providing mitochondria-targeted doxorubicin, and N. Varki and the UCSD histopathology core for preparation and analysis of mouse tissues. This work was supported by NIH grant no. R01 AR069876 and the Audrey Geisel Chair in Biomedical Science to G.S.S., NIH grant no. R01 CA216101 to G.S.S and S.M.K., NIH grant no. R35 CA197574 to P.M.G., NIH grant no. R01 CA237586 to Q.Y., NIH grant no. F31 AG062099 to A.G.S. and NIH grant no. P50 CA121974. A.P.W. was supported by grant no. RP170734 from the Cancer Prevention and Research Institute of Texas and grant no. W81XWH-17-1-0052 from the Office of the Assistant Secretary of Defense for Health Affairs, Peer Reviewed Medical Research Program. Z.W. was supported by the China Scholarship Counsel, K.C.M. by the Salk Excellerators Postdoctoral Fellowship and L.E.N. by the George E. Hewitt Foundation for Medical Research Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
Z.W. designed and performed experiments, analysed the data and wrote the paper. S.O. designed and performed experiments, analysed the data and wrote the paper. A.P.W. designed and performed experiments. A.G.S. performed experiments. X.Z. provided bioinformatics analyses. L.E.N. performed experiments. K.C.M. provided reagents and performed experiments. L.W. provided key reagents and performed experiments. P.L.S. designed and performed experiments. Y.L. performed experiments. V.T. performed experiments. M.B. and S.M.K. provided key reagents and funding. Q.Y. provided key reagents and funding. P.M.G. designed experiments and provided key reagents and funding. G.S.S. designed experiments, wrote the paper and provided funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
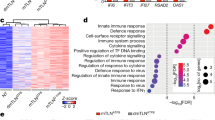
Extended Data Fig. 1 Additional analysis of innate immune signalling in MEFs.
a, Heat map of normalized expression values of the indicated ISGs (red font), interferon genes (black font), and NF-κB target genes (blue font) from our previously published microarray analysis4 of WT and Tfam+/- MEFs (two of each). b–d, RT-qPCR analyses of the indicated ISGs (b), NF-κB target genes (c) and interferon genes (d) in WT MEFs transfected with control (Ctrl) or Tfam siRNA for 72 h. e, Western blot probing the indicated NF-κB pathway proteins, TFAM and VDAC (loading control) in WT and Tfam+/- littermate MEFs. (n = 3 independent experiments.) f, RT-qPCR analysis of interferon β (Ifnb) gene expression in WT MEFs transfected with 2 µg poly(I:C) or lipofectamine only (Mock) for 9 h. g, RT-qPCR analysis of the indicated ISGs in WT MEFs cultured overnight in the presence of control media (plain), media conditioned by WT or Tfam+/- MEFs, or media conditioned by WT MEFs stimulated with poly(I:C) for 9 h (PIC-stimulated). The data shown are from one of two (f, g) or three (b–d) biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other biological replicates are provided as Supplementary Fig. 4. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, n.s. not significant (P > 0.05).
Extended Data Fig. 2 Additional analysis of the STAT1 null (Stat1-/-) condition in WT (Tfam+/+ Stat1+/+) and Tfam+/- MEFs.
a, Western blot of STAT1, TFAM and GAPDH (loading control) in littermate MEFs of the indicated Tfam and Stat1 genotypes (bottom) that were transfected with 2 µg of Poly(I:C) or lipofectamine only (Mock) for 12 h. (n = 3 independent experiments). MEFs described in a were analysed (all normalized to WT) for b, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers; c, mitochondrial mass using MitoTracker Green (MTG) and flow cytometry (mean fluorescence intensity, MFI, in arbitrary units, A.U., is plotted), and d, mitochondrial membrane potential using MitoTracker Deep Red (MTDR) and flow cytometry (MFI in A.U. is plotted). e, MEFs of the indicated genotypes were analysed by immunofluorescence for mitochondrial and mtDNA nucleoid morphology using antibodies against HSP60 (Mito., magenta) and DNA (DNA, cyan), respectively (n = 3 independent experiments). Images are Z-stack projections and scale bar represents 10 µm. f, RT-qPCR analysis of the ISGs Cxcl10 and Ifit1 in MEFs of the indicated genotypes. g, RT-qPCR analysis of the indicated ISGs in Tfam+/- MEFs transfected with control (Ctrl) or Irf3 siRNAs for 72 h. h, RT-qPCR analysis of the indicated ISGs in Tfam+/- Stat1-/- MEFs transfected with control (Ctrl) or Irf3 siRNAs for 72 h. i, Western blot showing siRNA knock-down of IRF9 or STAT2 in Tfam+/- MEFs compared to control (Ctrl) siRNA treated cells. GAPDH was probed as the loading control. (n = 3 independent experiments). j, RT-qPCR analysis of the indicated ISGs in Tfam+/- Stat1-/- MEFs transfected with control (Ctrl) or Irf9 siRNAs for 72 h. k, RT-qPCR analysis of the indicated ISGs in Tfam+/- Stat1-/- MEFs transfected with control (Ctrl) or Stat2 siRNAs for 72 h. c, d, error bars indicate mean ± s.d. of n = 3 biological replicates. For b the data shown are from one of two biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. For g, h, j and k, the data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other two biological replicates and the FACS gating strategy are provided as Supplementary Figs. 5 and 11. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001, n.s. not significant (P > 0.05).
Extended Data Fig. 3 Additional DNA damage and repair analysis in MEFs.
a, Pie charts showing the percentage of reported U-ISGF3-induced11 (green) and IRDS-related14 (blue) ISGs that are upregulated in Tfam+/- MEFs. The specific ISGs in these signatures are shown in the Table to the right, with the genes common to all three groups in bold. b-d, Stat1-/- and Tfam+/- Stat1-/- MEFs were treated with 1µM doxorubicin for 48 h followed by (b) Apoptosis analysis by flow cytometry using Annexin V and Propidium Iodide (PI); (Contour plots represent n=3 biological replicates). (c) Quantification of flow cytometry analysis in b for live cells (Annexin V and PI low), apoptotic cells (Annexin V high and PI low), and dead cells (Annexin V and PI high); (d) Cell viability analysis using the alamarBlue assay. c, d, Error bars indicate mean ± s.d. of (c) n=3 or (d) n=8 biological replicates. e, f, Analysis of nuclear DNA repair rate (that is the rate of γH2A.X and p-53BP1 foci resolution during recovery after 2 Gy IR). Nuclei are labelled with DAPI (blue), while γH2A.X (magenta), and p-53BP1 (green) were detected by immunofluorescence. Plotted to the right of the images is the average number of foci per nucleus at the indicated times. g and h, Analysis of the rate of induction of the nDNA damage response (that is the rate of γH2A.X and p-53BP1 foci formation) upon 2 Gy IR. Imaging was done as described in e and f. Plotted to the right is the percentage of positive cells (>20 γH2A.X foci per nucleus or >15 p-53BP1 foci per nucleus) at the indicated times. e–h, Scale bars represent 15 µm. e–h, Error bars indicate means ± s.d. of n=3 biological replicates, except for the 7.5 min time point in g, h, which are from n=2 biological replicates. In each replicate, 50 nuclei were quantified. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, n.s. not significant (P > 0.05).
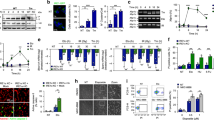
Extended Data Fig. 4 Additional data supporting the role of PARP9 in mtDNA-stress-mediated enhancement of nDNA damage responses.
a, Heat map of PARP family gene expression from previously published microarray data4 in WT and Tfam+/- littermate MEFs (two of each). b, RT-qPCR analysis of Parp9 expression in WT (Tfam+/+ Stat1+/+), Tfam+/-, Stat1-/- and Tfam+/- Stat1-/- littermate MEFs. c, RT-qPCR analysis of Parp9 expression at the indicated times after transfection with two Parp9 siRNAs (#1 and #2). b and c, the data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other two biological replicates are provided as Supplementary Figure 6. d, Tfam+/- MEFs were transfected with control (Ctrl) or one of two Parp9 siRNAs (#1 and #2) for 48 h and then assessed for cell viability using the alamarBlue assay after treatment with 1.0 or 1.5 µM doxorubicin (Dox) for 24 h. Error bars indicate mean ± s.d. of n=4 biological replicates. e–h, Analysis of DNA repair rate (that is the rate of γH2A.X and p-53BP1 foci resolution during recovery after (e, f) 2 Gy IR or (g, h) 12 h of 1µM doxorubicin-mediated damage. Nuclei are labelled with DAPI (blue), while γH2A.X (magenta), and p-53BP1 (green) were detected by immunofluorescence. (e–h) Plotted to the right of the images is the average number of foci per nucleus at the indicated times. Scale bars represent 15 µm. Error bars indicate means ± s.d. of n=3 biological replicates, with 50 nuclei quantified in each. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 5 Additional data supporting mtDNA stress-mediated ISG induction in MEFs.
a, Western blot probing STAT1, p-STAT1 (Y701), γH2A.X (DNA damage marker) and GAPDH (loading control) in WT MEFs treated with the indicated doses of doxorubicin (Dox) for 24 h. (n=3 independent experiments). b, RT-qPCR analysis of the indicated ISGs in WT (Stat1+/+) and Stat1 null (Stat1-/-) littermate MEFs challenged with (+Dox) or without 500 nM Dox for 24 h. c, WT MEFs treated with (Dox) or without (Ctrl) 1µM doxorubicin were analysed by immunofluorescence for mitochondrial and mtDNA nucleoid morphology by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan). (n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 µm. d and f, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in WT MEFs exposed to (d) 100 µM 2′−3′-dideoxycytidine (ddC) or (f) 450 ng/mL ethidium bromide (EtBr) for 10–12 days. e, g, RT-qPCR analysis of the ISGs Ifit1 and Ifit3 in (e) Control (Ctrl) and ddC-treated MEFs (described in d) challenged with (+Dox) or without (-Dox) 500nM doxorubicin for 16 h, and (g) Control (Ctrl) and EtBr-treated MEFs (described in f.) transfected with 2 µg dsDNA90 or lipofectamine only (Mock) for 9 h. h, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in LMTK- cells with (ρ+) or without (ρ˚) mtDNA. i, RT-qPCR analysis of the ISGs Ifit1 and Ifit3 in LMTK- ρ+ and ρ˚ cells (described in h.) challenged with 500 nM Dox for 24 h. j and k, MEFs were pre-treated with 100 µM chloramphenicol (Chlo) for 24 h followed by 500 nM Doxorubicin (Dox) treatment for 24 h. j, Western blot using an OXPHOS complex cocktail (mtDNA-encoded subunit is in red, nucleus-encoded subunits in black), γH2A.X (DNA damage marker) or actin (loading control) (n=3 independent experiments). k, RT-qPCR analysis of the ISGs Ifit1 and Ifit3. For e and g the data shown are from one of two biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. For b, d, f, h and i, the data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other biological replicates are provided as Supplementary Fig. 7. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 6 Doxorubicin promotes mtDNA stress-mediated ISG induction in MC-38 mouse colon cancer cells.
MC-38 cells treated with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 h and analysed by a, RT-qPCR analysis of the indicated ISGs and b, Western blot probing STAT1, p-STAT1 (Y701), γH2A.X (DNA damage marker) and GAPDH (loading control). (n=3 independent experiments) c, d, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers in WT MEFs exposed to (c) 100µM 2′−3′-dideoxycytidine (ddC) or (d) 200ng/mL ethidium bromide (EtBr) for 48 h. e, RT-qPCR analysis of the ISGs Ifit1 and Ifit3 in control (Ctrl) and ddC treated MC-38 cells (described in c) challenged with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 h. f, RT-qPCR analysis of the ISGs Ifit1 and Ifit3 in control (Ctrl) and EtBr-treated MC-38 cells (described in d) challenged with (+Dox) or without (-Dox) 150 nM doxorubicin for 24 h. g, RT-qPCR analysis of the indicated ISGs in MC-38 cells treated with 3 µM mitochondria-targeted doxorubicin (mitoDox) or DMSO (Mock) for 48 h. h, Western blot analysis in MC-38 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) gRNA control to determine the knockout efficiency. (n=1 only to validate) Vinculin was probed as the loading control. i–k, RT-qPCR analysis of the ISGs Ifit1 and Ifit3 in MC-38 cell pools transduced with the indicated gRNAs (described in h) that were transfected with (i) 2 µg dsDNA90 or (j) 2 µg Poly(I:C) (PIC) for 8 h or (k) challenged with 150 nM doxorubicin for 24 h. For i and j, the data shown are from one of two biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. For a, c-g and k, the data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other biological replicates are provided as Supplementary Fig. 8. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows: ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 7 TFAM deficiency induces mtDNA stress and DNA-damage resistance in MC-38 mouse colon cancer cells.
a-e, WT and TFAM deficient (TFD) MC-38 cells (see Methods section) were analysed. a, RT-qPCR analysis of Tfam mRNA. b, western blot probing STAT1 (L.E., long exposure; S.E., short exposure), TFAM and GAPDH (loading control) (n=3 independent experiments). c, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers. d, mitochondrial and mtDNA nucleoid morphology analyses by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan) (n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 µm. e, RT-qPCR analysis of the indicated ISGs. f, Western blot analysis in WT and TFD MC-38 cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) control gRNA to determine the knockout efficiency (n=1, only to validate). GAPDH was probed as the loading control. g, RT-qPCR analysis of the indicated ISGs in WT and TFD MC-38 cell pools transduced with the indicated gene-specific gRNAs (described in f). h, Cell survival analysis of WT and TFD MC-38 cells challenged with 500 nM doxorubicin for 48 h using the alamarBlue assay (n=7 biological replicates). a, c, e and g, the data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other two biological replicates are provided as Supplementary Fig. 9. All data were analysed with two-tailed unpaired student’s t tests. Asterisks indicate significance as follows:. ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Extended Data Fig. 8 Additional analysis of mtDNA stress and DNA-damage resistance in YUMMER mouse melanoma cells.
a, Western blot of STAT1, TFAM and GAPDH (loading control) in WT and Tfam deficient (TFD) YUMMER cells (n=3 independent experiments). b, Colony formation analysis of WT and TFD YUMMER cells challenged with 250nM doxorubicin (Dox) for 4 h. Plotted to the right is the quantification of relative cell survival (n=3 biological replicates). c, Western blot analysis in WT and TFD YUMMER cell pools transduced with the indicated gene-specific guide RNA (gRNA) or scrambled (Scr) control gRNA to determine the knockout efficiency (n=1, just to validate). GAPDH was probed as the loading control. d, Western blot of HA, TFAM and β-actin (Actin, loading control) in parental WT and TFD YUMMER cells, or WT and TFD YUMMER cells stably overexpressing empty vectors (EV) or HA-Flag-TFAM (OE) (n=3 independent experiments). e-g, YUMMER cells described in d were analysed as follows. e, mtDNA abundance (relative mtDNA copy number) by qPCR with D-loop primers. f, RT-qPCR analysis of the indicated ISGs. g, Mitochondrial and mtDNA nucleoid morphology analysis by immunofluorescence against HSP60 (Mito., magenta) and DNA (DNA, cyan)(n=3 independent experiments). Images are Z-stack projections and scale bar represents 10 µm. The data shown are from one of three biological replicates with the error bars indicating the mean ± s.d. of three technical replicates. The other two biological replicates are provided as Supplementary Fig. 10. ** P < 0.01, *** P < 0.001.
Extended Data Fig. 9 ISG expression is inversely correlates with mRNA expression of Tfam and other mtDNA-metabolism genes.
a-c, RNA-seq data from the CCLE23 (Cancer Cell Line Encyclopedia) was analysed. a, shown is the analysis of the upper and lower quartiles of Tfam RNA expression relative to the expression of the indicated ISGs, three interferon genes IFNA1, IFNB1 and IFNG (bold) and GAPDH (negative control). b, shown is the analysis of cells in the upper and lower quartiles of LIG3 (top row), NTHL1 (middle row), and POLG (bottom row) RNA expression relative to the expression of the indicated ISG. c, shown is the analysis of cells in the upper and lower quartiles of SOD2 (top row), SDHD (middle row) and VDAC1 (bottom row) RNA expression relative to the expression of the indicated ISGs. P values were calculated using Wilcoxon rank-sum test and displayed under each box plot.
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and Figs. 1–11
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 1
Unprocessed Western Blots
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Western Blots
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 2
Unprocessed Western Blots
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed Western Blots
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 6
Unprocessed Western Blots
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed Western Blots
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 8
Unprocessed Western Blots
Source Data Extended Data Fig. 9
Statistical Source Data
Rights and permissions
About this article
Cite this article
Wu, Z., Oeck, S., West, A.P. et al. Mitochondrial DNA stress signalling protects the nuclear genome. Nat Metab 1, 1209–1218 (2019). https://doi.org/10.1038/s42255-019-0150-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-019-0150-8
This article is cited by
-
Radiation-induced changes in energy metabolism result in mitochondrial dysfunction in salivary glands
Scientific Reports (2024)
-
Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal
Nature Cell Biology (2024)
-
Mitochondrial nucleic acids in innate immunity and beyond
Experimental & Molecular Medicine (2023)
-
Mitochondrial DNA-triggered innate immune response: mechanisms and diseases
Cellular & Molecular Immunology (2023)
-
Antecedent chromatin organization determines cGAS recruitment to ruptured micronuclei
Nature Communications (2023)