Abstract
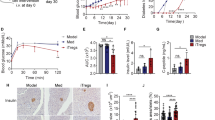
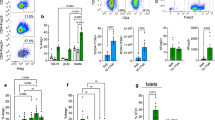
Type 1 diabetes (T1D) is characterized by pancreatic islet infiltration by autoreactive immune cells and a nearly complete loss of β cells1. Restoration of insulin-producing β cells coupled with immunomodulation to suppress the autoimmune attack has emerged as a potential approach to counter T1D2,3,4. Here we report that enhancing β-cell mass early in life, in two models of female non-obese diabetic (NOD) mice, results in immunomodulation of T cells, reduced islet infiltration and lower β-cell apoptosis, which together protect them from developing T1D. The animals displayed altered β-cell antigens; islet transplantation studies showed prolonged graft survival in the NOD-liver-specific insulin receptor knockout (LIRKO) model. Adoptive transfer of splenocytes from NOD-LIRKO mice prevented development of diabetes in prediabetic NOD mice. A substantial increase in the splenic CD4+CD25+Foxp3+ regulatory T cell (Treg) population was observed to underlie the protected phenotype since Treg-cell depletion rendered NOD-LIRKO mice diabetic. An increase in Treg cells coupled with activation of transforming growth factor-β/SMAD family member 3 signalling pathway in pathogenic T cells favoured reduced ability to kill β cells. These data support a previously unidentified observation that initiating β-cell proliferation, alone, before islet infiltration by immune cells alters the identity of β cells, decreases pathological self-reactivity of effector T cells and increases Treg cells to prevent the progression of T1D.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Transcriptomics data have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus under accession code GSE128315 for the murine microarray analysis. The mass spectrometry proteomics data reported in this paper have been deposited with ProteomeXchange (accession code: PXD013100) and MassIVE (accession code: MSV000083576). Further information on statistical parameters, software and study design can be found in the Nature Research Reporting Summary. The data that support the plots within this article and other findings of this study are available from the corresponding author upon reasonable request.
References
Mathis, D., Vence, L. & Benoist, C. β-Cell death during progression to diabetes. Nature 414, 792–798 (2001).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Tooley, J. E., Waldron-Lynch, F. & Herold, K. C. New and future immunomodulatory therapy in type 1 diabetes. Trends Mol. Med. 18, 173–181 (2012).
Johannesson, B., Sui, L., Freytes, D. O., Creusot, R. J. & Egli, D. Toward beta cell replacement for diabetes. EMBO J. 34, 841–855 (2015).
Michael, M. D. et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 (2000).
Chen, Y. G., Mathews, C. E. & Driver, J. P. The role of NOD mice in type 1 diabetes research: lessons from the past and recommendations for the future. Front. Endocrinol. (Lausanne) 9, 51 (2018).
Buchwald, P. et al. Comprehensive metabolomics study to assess longitudinal biochemical changes and potential early biomarkers in nonobese diabetic mice that progress to diabetes. J. Proteome Res. 16, 3873–3890 (2017).
Wilcox, N. S., Rui, J., Hebrok, M. & Herold, K. C. Life and death of β cells in type 1 diabetes: a comprehensive review. J. Autoimmun. 71, 51–58 (2016).
El Ouaamari, A. et al. SerpinB1 promotes pancreatic β cell proliferation. Cell Metab. 23, 194–205 (2016).
Stadinski, B. D. et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat. Immunol. 11, 225–231 (2010).
Delong, T. et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 (2016).
Schuster, C. et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity 42, 942–952 (2015).
Singhal, G. et al. Fibroblast growth factor 21 (FGF21) protects against high fat diet induced inflammation and islet hyperplasia in pancreas. PLoS ONE 11, e0148252 (2016).
Sun, M. Y. et al. Autofluorescence imaging of living pancreatic islets reveals fibroblast growth factor-21 (FGF21)-induced metabolism. Biophys. J. 103, 2379–2388 (2012).
Wicker, L. S. et al. β2-Microglobulin-deficient NOD mice do not develop insulitis or diabetes. Diabetes 43, 500–504 (1994).
Kay, T. W., Parker, J. L., Stephens, L. A., Thomas, H. E. & Allison, J. RIP-beta 2-microglobulin transgene expression restores insulitis, but not diabetes, in beta 2-microglobulin null nonobese diabetic mice. J. Immunol. 157, 3688–3693 (1996).
Tang, Q. & Bluestone, J. A. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat. Immunol. 9, 239–244 (2008).
Zhang, Y., Bandala-Sanchez, E. & Harrison, L. C. Revisiting regulatory T cells in type 1 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 19, 271–278 (2012).
Tang, J. et al. IL-25 promotes the function of CD4+CD25+ T regulatory cells and prolongs skin-graft survival in murine models. Int. Immunopharmacol. 28, 931–937 (2015).
Shultz, L. D. et al. Humanized NOD/LtSz-scid IL2 receptor common gamma chain knockout mice in diabetes research. Ann. N. Y. Acad. Sci. 1103, 77–89 (2007).
Tsai, S., Shameli, A. & Santamaria, P. CD8+ T cells in type 1 diabetes. Adv. Immunol. 100, 79–124 (2008).
Dirice, E. et al. Soluble factors secreted by T cells promote β-cell proliferation. Diabetes 63, 188–202 (2014).
McKarns, S. C. & Schwartz, R. H. Distinct effects of TGF-β1 on CD4+ and CD8+ T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. J. Immunol. 174, 2071–2083 (2005).
Delisle, J. S. et al. The TGF-β-Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes Immun. 14, 115–126 (2013).
Park, B. V. et al. TGFβ1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in cancer. Cancer Discov. 6, 1366–1381 (2016).
Wang, H. et al. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2, a002279 (2010).
Hara, T. et al. Biglycan intensifies ALK5-Smad2/3 signaling by TGF-β1 and downregulates syndecan-4 in cultured vascular endothelial cells. J. Cell. Biochem. 118, 1087–1096 (2017).
Tzachanis, D. et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182 (2001).
Tzachanis, D. & Boussiotis, V. A. Tob, a member of the APRO family, regulates immunological quiescence and tumor suppression. Cell Cycle 8, 1019–1025 (2009).
Adurthi, S. et al. Oestrogen receptor-α binds the FOXP3 promoter and modulates regulatory T-cell function in human cervical cancer. Sci. Rep. 7, 17289 (2017).
Polanczyk, M. J. et al. Cutting edge: estrogen drives expansion of the CD4+CD25+ regulatory T cell compartment. J. Immunol. 173, 2227–2230 (2004).
Skyler, J. S. Hope vs hype: where are we in type 1 diabetes? Diabetologia 61, 509–516 (2018).
Thorn, L. M. et al. Effect of parental type 2 diabetes on offspring with type 1 diabetes. Diabetes Care 32, 63–68 (2009).
Zalloua, P. A. et al. Type-2 diabetes family history delays the onset of type-1 diabetes. J. Clin. Endocrinol. Metab. 87, 3192–3196 (2002).
Warram, J. H., Krolewski, A. S., Gottlieb, M. S. & Kahn, C. R. Differences in risk of insulin-dependent diabetes in offspring of diabetic mothers and diabetic fathers. N. Engl. J. Med. 311, 149–152 (1984).
Kulkarni, R. N. et al. Altered function of insulin receptor substrate-1-deficient mouse islets and cultured β-cell lines. J. Clin. Invest. 104, R69–R75 (1999).
Dirice, E. et al. Inhibition of DYRK1A stimulates human β-cell proliferation. Diabetes 65, 1660–1671 (2016).
Flodström-Tullberg, M. et al. Target cell expression of suppressor of cytokine signaling-1 prevents diabetes in the NOD mouse. Diabetes 52, 2696–2700 (2003).
King, A. J. et al. Normal relationship of β- and non-β-cells not needed for successful islet transplantation. Diabetes 56, 2312–2318 (2007).
Huang, E. L. et al. SNaPP: Simplified Nanoproteomics Platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology 157, 1307–1314 (2016).
Clair, G. et al. Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci. Rep. 6, 39223 (2016).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Delong, T. et al. Islet amyloid polypeptide is a target antigen for diabetogenic CD4+ T cells. Diabetes 60, 2325–2330 (2011).
Pauken, K. E. et al. Cutting edge: type 1 diabetes occurs despite robust anergy among endogenous insulin-specific CD4 T cells in NOD mice. J. Immunol. 191, 4913–4917 (2013).
Pauken, K. E. et al. Cutting edge: identification of autoreactive CD4+ and CD8+ T cell subsets resistant to PD-1 pathway blockade. J. Immunol. 194, 3551–3555 (2015).
Shi, W., Oshlack, A. & Smyth, G. K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 38, e204 (2010).
Ritchie, M. E. et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 7, 261 (2006).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Herwig, R., Hardt, C., Lienhard, M. & Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 11, 1889–1907 (2016).
Acknowledgements
We thank the late A. Rossini for discussions during the early stages of this project. We thank C. R. Kahn for discussions and sharing the LIRKO model. We thank D. Mathis, J. Gaglia, C. Mathews, G. C. Weir and F. Bosch for discussions regarding various aspects of the studies and the NOD-RAG1−/− mice. We thank H. Davidson for kindly providing the anti-ZnT8 (C-term) and anti-Phogrin (C-term) sera and F. M. Jarvis for assistance with the E2 assays. We thank C. Cahill for assistance with confocal microscopy, L. Kannan, Z. Fu and G. Sankaranarayanan for the ELISAs, J. Hollister-Lock for assistance with the mouse islet isolations and J. Dreyfuss and H. Pan for assistance in the data analyses. The proteomics experiments were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the United States Department of Energy (DOE) and located at the Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under contract no. DE-AC05-76RL0 1830. E.D. was supported by a Senior Juvenile Diabetes Research Foundation Fellowship (no. JDRF-3-APF-2014-220-A-N). This project was funded by JDRF grant no. 1-SRA-355-Q-R (to R.N.K.) and in part from National Institutes of Health (NIH) grant no. RO1 067536 (to R.N.K.), NIH grant no. RO1 103215 (to R.N.K.) and grant no. UC4 DK104167 (to W.J.Q. and R.N.K.). R.N.K. acknowledges support from the Margaret A. Congleton Chair, Joslin Diabetes and Endocrinology Research Center (grant no. P30 386836). R.D.S. was supported by grant no. P41 GM103493. B.T.F. was supported by grant no. R01 AI106791. R.L.B. was supported by grant no. R21 AI133059 and an ADA Junior Faculty Award no. 1-15-JF-04. K.H. was supported by grant no. R01 DK081166.
Author information
Authors and Affiliations
Contributions
E.D. conceived the idea, designed and performed the experiments, analysed the data and wrote the manuscript. A.E., S.Kahraman, D.F.J., G.B., B.Y., R.W.S.N., H.V. and A.K.K.T. researched the data, provided technical support and/or critical discussions of the manuscript. C.S. performed the bone marrow experiments. M.K. and Y.I. assisted with FACS analysis. J.H. performed the immunohistochemistry. P.D.P., R.D.S. and W.J.Q. performed the proteomics experiments and data analysis. A.J.D., T.M. and B.T.F performed the tetramer assay and analysed the data. R.L.B. and K.H. performed the antigen assay and analysed the data. T.S. and S. Kissler contributed to discussions and the experiments on immune regulation. R.N.K. conceived the idea, designed the experiments, supervised the project and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Information
Supplementary Figs. 1–12
Supplementary Table 1
Selected pathways upregulated in 8 week CD8+ T cells.
Supplementary Table 2
Selected pathways upregulated in 12 week CD8+ T cells.
Supplementary Table 3
Selected pathways downregulated in 12 week CD8+ T cells.
Supplementary Table 4
Selected pathways upregulated in 8 week Treg cells.
Supplementary Table 5
Selected pathways downregulated in 12 week Treg cells.
Supplementary Table 6
Selected pathways upregulated in 12 week Treg cells.
Rights and permissions
About this article
Cite this article
Dirice, E., Kahraman, S., De Jesus, D.F. et al. Increased β-cell proliferation before immune cell invasion prevents progression of type 1 diabetes. Nat Metab 1, 509–518 (2019). https://doi.org/10.1038/s42255-019-0061-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-019-0061-8
This article is cited by
-
Redox regulation of m6A methyltransferase METTL3 in β-cells controls the innate immune response in type 1 diabetes
Nature Cell Biology (2024)
-
Talin-1 inhibits Smurf1-mediated Stat3 degradation to modulate β-cell proliferation and mass in mice
Cell Death & Disease (2023)
-
Islet autoimmunity in human type 1 diabetes: initiation and progression from the perspective of the beta cell
Diabetologia (2023)
-
Activation of arcuate nucleus glucagon-like peptide-1 receptor-expressing neurons suppresses food intake
Cell & Bioscience (2022)
-
Interactions between islets and regulatory immune cells in health and type 1 diabetes
Diabetologia (2021)