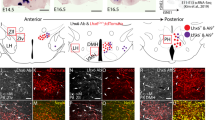
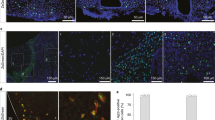
Abstract
Heterogeneous populations of hypothalamic neurons orchestrate energy balance via the release of specific signatures of neuropeptides. However, how specific intracellular machinery controls peptidergic identities and function of individual hypothalamic neurons remains largely unknown. The transcription factor T-box 3 (Tbx3) is expressed in hypothalamic neurons sensing and governing energy status, whereas human TBX3 haploinsufficiency has been linked with obesity. Here, we demonstrate that loss of Tbx3 function in hypothalamic neurons causes weight gain and other metabolic disturbances by disrupting both the peptidergic identity and plasticity of Pomc/Cart and Agrp/Npy neurons. These alterations are observed after loss of Tbx3 in both immature hypothalamic neurons and terminally differentiated mouse neurons. We further establish the importance of Tbx3 for body weight regulation in Drosophila melanogaster and show that TBX3 is implicated in the differentiation of human embryonic stem cells into hypothalamic Pomc neurons. Our data indicate that Tbx3 directs the terminal specification of neurons as functional components of the melanocortin system and is required for maintaining their peptidergic identity. In summary, we report the discovery of a key mechanistic process underlying the functional heterogeneity of hypothalamic neurons governing body weight and systemic metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. The RNA-seq database generated in our paper has been made publicly available through Gene Expression Omnibus (GEO accession number GSE119883).
References
Cone, R. D. Anatomy and regulation of the central melanocortin system. Nat. Neurosci. 8, 571–578 (2005).
Gautron, L., Elmquist, J. K. & Williams, K. W. Neural control of energy balance: translating circuits to therapies. Cell 161, 133–145 (2015).
Koch, M. & Horvath, T. L. Molecular and cellular regulation of hypothalamic melanocortin neurons controlling food intake and energy metabolism. Mol. Psychiatry 19, 752–761 (2014).
Morton, G. J., Meek, T. H. & Schwartz, M. W. Neurobiology of food intake in health and disease. Nat. Rev. Neurosci. 15, 367–378 (2014).
Knight, Z. A. et al. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137 (2012).
Allison, M. B. et al. TRAP-seq defines markers for novel populations of hypothalamic and brainstem LepRb neurons. Mol. Metab. 4, 299–309 (2015).
Campbell, J. N. et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017).
Wansleben, S., Peres, J., Hare, S., Goding, C. R. & Prince, S. T-box transcription factors in cancer biology. Biochim. Biophys. Acta 1846, 380–391 (2014).
Ang, L. T. et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 22, 2190–2205 (2018).
Suzuki, A., Sekiya, S., Büscher, D., Izpisúa Belmonte, J. C. & Taniguchi, H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development 135, 1589–1595 (2008).
Weidgang, C. E. et al. TBX3 Directs cell-fate decision toward mesendoderm. Stem Cell Rep. 1, 248–265 (2013).
Eriksson, K. S. & Mignot, E. T-box 3 is expressed in the adult mouse hypothalamus and medulla. Brain Res. 1302, 233–239 (2009).
Linden, H., Williams, R., King, J., Blair, E. & Kini, U. Ulnar mammary syndrome and TBX3: expanding the phenotype. Am. J. Med. Genet. A 149A, 2809–2812 (2009).
Schinzel, A. The ulnar-mammary syndrome: an autosomal dominant pleiotropic gene. Clin. Genet. 32, 160–168 (1987).
Grill, H. J. & Hayes, M. R. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell. Metab. 16, 296–309 (2012).
Joly-Amado, A. et al. The hypothalamic arcuate nucleus and the control of peripheral substrates. Best Pract. Res. Clin. Endocrinol. Metab. 28, 725–737 (2014).
Clasadonte, J. & Prevot, V. The special relationship: glia-neuron interactions in the neuroendocrine hypothalamus. Nat. Rev. Endocrinol. 14, 25–44 (2018).
Pontecorvi, M., Goding, C. R., Richardson, W. D. & Kessaris, N. Expression of Tbx2 and Tbx3 in the developing hypothalamic-pituitary axis. Gene. Expr. Patterns 8, 411–417 (2008).
Tschöp, M. H. et al. A guide to analysis of mouse energy metabolism. Nat. Methods 9, 57–63 (2011).
Tong, Q., Ye, C.-P., Jones, J. E., Elmquist, J. K. & Lowell, B. B. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat. Neurosci. 11, 998–1000 (2008).
Balthasar, N. et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron 42, 983–991 (2004).
Kumar, P. P. et al. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. eLife 3, e02805 (2014).
Coll, M., Seidman, J. G. & Müller, C. W. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10, 343–356 (2002).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Bandyopadhyay, S. et al. A human map kinase interactome. Nat. Methods 7, 801–805 (2010).
Padilla, S. L., Carmody, J. S. & Zeltser, L. M. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat. Med. 16, 403–405 (2010).
Toda, C., Santoro, A., Kim, J. D. & Diano, S. POMC neurons: from birth to death. Annu. Rev. Physiol. 79, 209–236 (2017).
Hahn, T. M., Breininger, J. F., Baskin, D. G. & Schwartz, M. W. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat. Neurosci. 1, 271–272 (1998).
Sousa-Ferreira, L., de Almeida, L. P. & Cavadas, C. Role of hypothalamic neurogenesis in feeding regulation. Trends Endocrinol. Metab. 25, 80–88 (2014).
Mizuno, T. M. et al. Hypothalamic pro-opiomelanocortin mrna is reduced by fasting in ob/ob and db/db mice, but is stimulated by leptin. Diabetes 47, 294–297 (1998).
Wilson, V. & Conlon, F. L. The T-box family. Genome Biol. 3, REVIEWS3008 (2002).
Wang, L. et al. Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J. Clin. Invest. 125, 796–808 (2015).
Wang, L., Egli, D. & Leibel, R. L. Efficient generation of hypothalamic neurons from human pluripotent stem cells. Curr. Protoc. Hum. Genet. 90, 21.5.1–21.5.14 (2016).
Wang, L. et al. PC1/3 deficiency impacts pro-opiomelanocortin processing in human embryonic stem cell-derived hypothalamic neurons. Stem Cell Rep. 8, 264–277 (2017).
Coupe, B. & Bouret, S. G. Development of the hypothalamic melanocortin system. Front Endocrinol. (Lausanne) 4, 38 (2013).
Pelling, M. et al. Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev. Biol. 349, 406–416 (2011).
Lee, B. et al. Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nat. Commun. 9, 2026 (2018).
Nasif, S. et al. Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proc. Natl Acad. Sci. USA 112, E1861–E1870 (2015).
Lee, B., Lee, S., Lee, S.-K. & Lee, J. W. The LIM-homeobox transcription factor Isl1 plays crucial roles in the development of multiple arcuate nucleus neurons. Development 143, 3763–3773 (2016).
Sakkou, M. et al. A role for brain-specific homeobox factor Bsx in the control of hyperphagia and locomotory behavior. Cell Metab. 5, 450–463 (2007).
Messina, A. et al. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat. Neurosci. 19, 835–844 (2016).
Greenman, Y. et al. Postnatal ablation of POMC neurons induces an obese phenotype characterized by decreased food intake and enhanced anxiety-like behavior. Mol. Endocrinol. 27, 1091–1102 (2013).
Morton, G. J. & Schwartz, M. W. The NPY/AgRP neuron and energy homeostasis. Int. J. Obes. Relat. Metab. Disord. 25 (Suppl. 5), S56–S62 (2001).
Luquet, S., Perez, F. A., Hnasko, T. S. & Palmiter, R. D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science 310, 683–685 (2005).
Tan, K., Knight, Z. A. & Friedman, J. M. Ablation of AgRP neurons impairs adaption to restricted feeding. Mol. Metab. 3, 694–704 (2014).
Bouret, S. G. & Simerly, R. B. Minireview: leptin and development of hypothalamic feeding circuits. Endocrinology 145, 2621–2626 (2004).
Zhan, C. et al. Acute and long-term suppression of feeding behavior by pomc neurons in the brainstem and hypothalamus, respectively. J. Neurosci. 33, 3624–3632 (2013).
Nogueiras, R. et al. The central melanocortin system directly controls peripheral lipid metabolism. J. Clin. Invest. 117, 3475–3488 (2007).
Burbridge, S., Stewart, I. & Placzek, M. Development of the neuroendocrine hypothalamus. Compr. Physiol. 6, 623–643 (2016).
Dulcis, D., Jamshidi, P., Leutgeb, S. & Spitzer, N. C. Neurotransmitter switching in the adult brain regulates behavior. Science 340, 449–453 (2013).
Gascón, S., Masserdotti, G., Russo, G. L. & Götz, M. Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21, 18–34 (2017).
Frank, D. U., Emechebe, U., Thomas, K. R. & Moon, A. M. Mouse Tbx3 mutants suggest novel molecular mechanisms for ulnar-mammary syndrome. PLoS One 8, e67841 (2013).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
van den Pol, A. N. et al. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J. Neurosci. 29, 4622–4639 (2009).
Cowley, M. A. et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411, 480–484 (2001).
Monory, K. et al. The endocannabinoid system controls key epileptogenic circuits in the hippocampus. Neuron 51, 455–466 (2006).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 13, 2513–2526 (2014).
Marco-Sola, S., Sammeth, M., Guigó, R. & Ribeca, P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat. Methods 9, 1185–1188 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Leek, J. T. & Storey, J. D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, e161 (2007).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25–29 (2000).
Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl Acad. Sci. USA 98, 12596–12601 (2001).
Shen, J., Dorner, C., Bahlo, A. & Pflugfelder, G. O. optomotor-blind suppresses instability at the A/P compartment boundary of the Drosophila wing. Mech. Dev. 125, 233–246 (2008).
Hildebrandt, A., Bickmeyer, I. & Kühnlein, R. P. Reliable Drosophila body fat quantification by a coupled colorimetric assay. PLoS One 6, e23796 (2011).
Gáliková, M., Klepsatel, P., Xu, Y. & Kühnlein, R. P. The obesity-related adipokinetic hormone controls feeding and expression of neuropeptide regulators of Drosophila metabolism. Eur. J. Lipid Sci. Tech. 119, 1600138 (2017).
Klepsatel, P., Gáliková, M., Xu, Y. & Kühnlein, R. P. Thermal stress depletes energy reserves in Drosophila. Sci. Rep. 6, 33667 (2016).
Mayer, L. R., Diegelmann, S., Abassi, Y., Eichinger, F. & Pflugfelder, G. O. Enhancer trap infidelity in Drosophila optomotor-blind. Fly 7, 118–128 (2013).
Baumbach, J., Xu, Y., Hehlert, P. & Kühnlein, R. P. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J. Genet. Genom. 41, 283–292 (2014).
Shen, J., Dahmann, C. & Pflugfelder, G. O. Spatial discontinuity of optomotor-blind expression in the Drosophila wing imaginal disc disrupts epithelial architecture and promotes cell sorting. BMC Dev. Biol. 10, 23 (2010).
Stratigopoulos, G., De Rosa, M. C., LeDuc, C. A., Leibel, R. L. & Doege, C. A. DMSO increases efficiency of genome editing at two non-coding loci. PLoS One 13, e0198637 (2018).
Santos, D. P., Kiskinis, E., Eggan, K. & Merkle, F. T. Comprehensive protocols for crispr/cas9-based gene editing in human pluripotentstem cells. Curr. Protoc. Stem Cell Biol. 38, 5B.6.1–5B.6.60 (2016).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Crawford, T. Q. & Roelink, H. The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dyn. 236, 886–892 (2007).
Nelson, B. R., Hartman, B. H., Georgi, S. A., Lan, M. S. & Reh, T. A. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 304, 479–498 (2007).
Trowe, M.-O. et al. Inhibition of Sox2-dependent activation of Shh in the ventral diencephalon by Tbx3 is required for formation of the neurohypophysis. Development 140, 2299–2309 (2013).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Acknowledgements
We thank A. Kispert and M.-O. Trowe (Institut für Molekularbiologie, Medizinische Hochschule Hannover, Hannover, Germany) for kindly providing Tbx3-deficient embryos, J. Friedman (The Rockefeller University, Howard Hughes Medical Institute, New York, NY, USA) for scientific guidance and for graciously providing access to data shown in ref. 5, M. Guzmán (Complutense University, Madrid, Spain) for assistance with the generation of AAV-GFP and AAV-Cre viral particles, the Bloomington Drosophila Stock Center (BDSC) (NIH P40OD018537) for fly stocks, and C. Layritz, H. Hoffmann, N. Wiegert and C. L. Holleman for technical assistance and assistance with animal studies. A.F. is supported by a postoctoral fellowship from the Canadian Institutes of Health Research (Funding reference no. 152588). V.V.T. is supported by NIH-NIDDK grant 5K23DK110539 and in part by the Baylor-Hopkins Center for Mendelian Genomics through NHGRI grant 5U54HG006542. C.A.D. is supported by funding from the NIH (R01 DK52431, R01 DK110113 and P30 DK26687) and Columbia Stem Cell Initiative Seed Fund Program. We thank the Fondation Recherche Medicale (ARF20140129235, L.B.). This work was strongly supported by the Helmholtz Alliance ICEMED & the Helmholtz Initiative on Personalized Medicine iMed by Helmholtz Association. This work was supported in part by the Helmholtz cross-program topic ‘Metabolic Dysfunction’, the European Research Council ERC (AdG HypoFlam no. 695054) and in part by funding to M.H.T., Y.L., B.L. and V.K. from the Alexander von Humboldt Foundation.
Author information
Authors and Affiliations
Contributions
C.Q. and A.F. designed and performed the experiments and interpreted the data. Y.X., G.C., B.L. Y.-T.T., A.R., M.W., M.C.D., V.K., R.R., V.V.T., E.G., T.M.S., A.-L.P., T.G., O.L., A.C.-S., D.K., L.B., S.C.W., G.O.P., R.N., L.Z., I.C.G.K., A.M., C.G.-C., M.M., M.T. and C.A.D. performed experiments and/or edited the manuscript. M.H.T. conceptualized the project, interpreted the data, and cowrote the manuscript together with C.Q. and A.F.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–6
Supplementary Table 1
qRT–PCR primer sequences
Supplementary Table 2
Reference proteins and proteins enriched with Tbx3 immunoprecipitation
Supplementary Table 3
Lineage tracing and neuropeptide expression.
Supplementary Table 4
Full vector sequence of pCas9_CD4
Rights and permissions
About this article
Cite this article
Quarta, C., Fisette, A., Xu, Y. et al. Functional identity of hypothalamic melanocortin neurons depends on Tbx3. Nat Metab 1, 222–235 (2019). https://doi.org/10.1038/s42255-018-0028-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-018-0028-1
This article is cited by
-
TBX3 is dynamically expressed in pancreatic organogenesis and fine-tunes regeneration
BMC Biology (2023)
-
Ontogenetic rules for the molecular diversification of hypothalamic neurons
Nature Reviews Neuroscience (2022)
-
Developmental programming of hypothalamic melanocortin circuits
Experimental & Molecular Medicine (2022)
-
POMC neuronal heterogeneity in energy balance and beyond: an integrated view
Nature Metabolism (2021)
-
Murine neuronatin deficiency is associated with a hypervariable food intake and bimodal obesity
Scientific Reports (2021)