Abstract
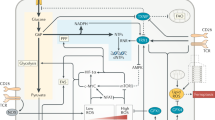
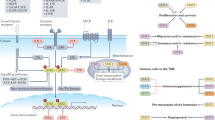
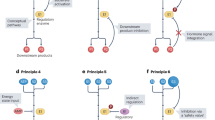
Metabolic reprogramming has become a key focus for both immunologists and cancer biologists, with exciting advances providing new insights into the mechanisms underlying disease. There is now extensive evidence that intermediates and derivatives of the mitochondrial Krebs cycle—metabolites traditionally associated with bioenergetics or biosynthesis—also possess ‘non-metabolic’ signalling functions. In this review, we summarize the non-metabolic signalling mechanisms of succinate, fumarate, itaconate, 2-hydroxyglutarate isomers (d-2-hydroxyglutarate and l-2-hydroxyglutarate) and acetyl-CoA, with a specific focus on how such signalling pathways alter immune cell and transformed cell function. We believe that the insights gained from immune and cancer cells that are summarized here will also be useful for understanding and treating a range of other diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
He, W. et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature 429, 188–193 (2004).
Chouchani, E. T. et al. A unifying mechanism for mitochondrial superoxide production during ischemia–reperfusion injury. Cell. Metab. 23, 254–263 (2016).
Rubic, T. et al. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat. Immunol. 9, 1261–1269 (2008).
Lei, W. et al. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl Acad. Sci. USA. 115, 5552–5557 (2018).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
Schneider, C. et al. A metabolite-triggered tuft cell–ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284.e14 (2018).
Littlewood-Evans, A. et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J. Exp. Med. 213, 1655–1662 (2016).
Kim, S. et al. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One 9, e97501 (2014).
Hollander, A. P., Corke, K. P., Freemont, A. J. & Lewis, C. E. Expression of hypoxia-inducible factor 1α by macrophages in the rheumatoid synovium: implications for targeting of therapeutic genes to the inflamed joint. Arthritis Rheum. 44, 1540–1544 (2001).
Sadagopan, N. et al. Circulating succinate is elevated in rodent models of hypertension and metabolic disease. Am. J. Hypertens. 20, 1209–1215 (2007).
van Diepen, J. A. et al. SUCNR1-mediated chemotaxis of macrophages aggravates obesity-induced inflammation and diabetes. Diabetologia 60, 1304–1313 (2017).
Mills, E. L. et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 560, 102–106 (2018).
Lewis, G. D. et al. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2, 33ra37 (2010).
Peruzzotti-Jametti, L. et al. Macrophage-derived extracellular succinate licenses neural stem cells to suppress chronic neuroinflammation. Cell Stem Cell 22, 355–368.e13 (2018).
Bhuniya, D. et al. Discovery of a potent and selective small molecule hGPR91 antagonist. Bioorg. Med. Chem. Lett. 21, 3596–3602 (2011).
Geubelle, P. et al. Identification and pharmacological characterization of succinate receptor agonists. Br. J. Pharmacol. 174, 796–808 (2017).
Tannahill, G. M. & O’Neill, L. A. The emerging role of metabolic regulation in the functioning of Toll-like receptors and the NOD-like receptor Nlrp3. FEBS Lett. 585, 1568–1572 (2011).
Semenza, G. L. & Wang, G. L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 (1992).
Semenza, G. L. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 24, 97–106 (2009).
Epstein, A. C. R. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Maxwell, P. H. et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 (1999).
Pearce, E. L. & Pearce, E. J. Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013).
Frede, S., Stockmann, C., Freitag, P. & Fandrey, J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-κB. Biochem. J. 396, 517–527 (2006).
Wu, W. & Zhao, S. Metabolic changes in cancer: beyond the Warburg effect. Acta Biochim. Biophys. Sin. (Shanghai) 45, 18–26 (2013).
Selak, M. A. et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 7, 77–85 (2005).
Rasola, A., Neckers, L. & Picard, D. Mitochondrial oxidative phosphorylation TRAP(1)ped in tumor cells. Trends Cell Biol. 24, 455–463 (2014).
Mu, X. et al. Oncometabolite succinate promotes angiogenesis by upregulating VEGF expression through GPR91-mediated STAT3 and ERK activation. Oncotarget 8, 13174–13185 (2017).
Guzy, R. D., Sharma, B., Bell, E., Chandel, N. S. & Schumacker, P. T. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 28, 718–731 (2008).
Hagen, T. Oxygen versus reactive oxygen in the regulation of HIF-1α: the balance tips. Biochem. Res. Int. 2012, 436981 (2012).
Folbergrová, J., Ljunggren, B., Norberg, K. & Siesjö, B. K. Influence of complete ischemia on glycolytic metabolites, citric acid cycle intermediates, and associated amino acids in the rat cerebral cortex. Brain Res. 80, 265–279 (1974).
Chouchani, E. T. et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 (2014).
Zhang, W. et al. Evidence that hypoxia-inducible factor-1 (HIF-1) mediates transcriptional activation of interleukin-1β (IL-1β) in astrocyte cultures. J. Neuroimmunol. 174, 63–73 (2006).
Peyssonnaux, C. et al. Cutting edge: essential role of hypoxia inducible factor-1α in development of lipopolysaccharide-induced sepsis. J. Immunol. 178, 7516–7519 (2007).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470.e13 (2016).
El-Khoury, R. et al. Engineering the alternative oxidase gene to better understand and counteract mitochondrial defects: state of the art and perspectives. Br. J. Pharmacol. 171, 2243–2249 (2014).
Jin, Z., Wei, W., Yang, M., Du, Y. & Wan, Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell. Metab. 20, 483–498 (2014).
Murphy, M. P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009).
Bénit, P. et al. Unsuspected task for an old team: succinate, fumarate and other Krebs cycle acids in metabolic remodeling. Biochim. Biophys. Acta 1837, 1330–1337 (2014).
Brigati, C. et al. Inflammation, HIF-1, and the epigenetics that follows. Mediators Inflamm. 2010, 263914 (2010).
Benn, C. S., Netea, M. G., Selin, L. K. & Aaby, P. A small jab—a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. 34, 431–439 (2013).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Cheng, S. C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Xie, L. et al. First succinyl-proteome profiling of extensively drug-resistant Mycobacterium tuberculosis revealed involvement of succinylation in cellular physiology. J. Proteome Res. 14, 107–119 (2015).
Feng, S. et al. Succinyl-proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway. BMC Genomics 18, 598 (2017).
He, D. et al. Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J. Proteome Res. 15, 879–890 (2016).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 (2012).
Du, J. et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Gibson, G. E. et al. α-ketoglutarate dehydrogenase complex–dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 134, 86–96 (2015).
Wang, F. et al. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1β production and to prevent dss-induced colitis in mice. Cell Rep. 19, 2331–2344 (2017).
Cordes, T. et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 (2016).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Meiser, J. et al. Pro-inflammatory macrophages sustain pyruvate oxidation through pyruvate dehydrogenase for the synthesis of itaconate and to enable cytokine expression. J. Biol. Chem. 291, 3932–3946 (2016).
Németh, B. et al. Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. FASEB J. 30, 286–300 (2016).
Okabe, M., Lies, D., Kanamasa, S. & Park, E. Y. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl. Microbiol. Biotechnol. 84, 597–606 (2009).
Lee, C. G., Jenkins, N. A., Gilbert, D. J., Copeland, N. G. & O’Brien, W. E. Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. Immunogenetics 41, 263–270 (1995).
Shin, J. H. et al. 1H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J. Proteome Res. 10, 2238–2247 (2011).
Sugimoto, M. S. H. Y. et al. Non-targeted metabolite profiling in activated macrophage secretion. Metabolomics 8, 624–633 (2011).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA. 110, 7820–7825 (2013).
McFadden, B. A. & Purohit, S. Itaconate, an isocitrate lyase-directed inhibitor in Pseudomonas indigofera. J. Bacteriol. 131, 136–144 (1977).
Naujoks, J. et al. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 12, e1005408 (2016).
Williams, J. O., Roche, T. E. & McFadden, B. A. Mechanism of action of isocitrate lyase from Pseudomonas indigofera. Biochemistry 10, 1384–1390 (1971).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBξ–ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Nair, S. et al. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med. 215, 1035–1045 (2018).
Ackermann, W. W. & Potter, V. R. Enzyme inhibition in relation to chemotherapy. Proc. Soc. Exp. Biol. Med. 72, 1–9 (1949).
El Azzouny, M. et al. Dimethyl itaconate is not metabolized into itaconate intracellularly. J. Biol. Chem. 292, 4766–4769 (2017).
Kobayashi, E. H. et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 7, 11624 (2016).
Brennan, M. S. et al. Dimethyl fumarate and monoethyl fumarate exhibit differential effects on KEAP1, NRF2 activation, and glutathione depletion in vitro. PLoS One 10, e0120254 (2015).
Saito, R. et al. Characterizations of three major cysteine sensors of Keap1 in stress response. Mol. Cell Biol. 36, 271–284 (2015).
Li, Y. et al. Immune responsive gene 1 (IRG1) promotes endotoxin tolerance by increasing A20 expression in macrophages through reactive oxygen species. J. Biol. Chem. 288, 16225–16234 (2013).
Van Quickelberghe, E. et al. Identification of immune-responsive gene 1 (IRG1) as a target of A20. J. Proteome Res. 17, 2182–2191 (2018).
Jamal Uddin, M. et al. IRG1 induced by heme oxygenase-1/carbon monoxide inhibits LPS-mediated sepsis and pro-inflammatory cytokine production. Cell Mol. Immunol. 13, 170–179 (2016).
Cheon, Y. P., Xu, X., Bagchi, M. K. & Bagchi, I. C. Immune-responsive gene 1 is a novel target of progesterone receptor and plays a critical role during implantation in the mouse. Endocrinology 144, 5623–5630 (2003).
Luan, H. H. & Medzhitov, R. Food fight: role of itaconate and other metabolites in antimicrobial defense. Cell. Metab. 24, 379–387 (2016).
Sherwin, J. R. et al. Identification of genes regulated by leukemia-inhibitory factor in the mouse uterus at the time of implantation. Mol. Endocrinol. 18, 2185–2195 (2004).
Weiss, J. M. et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 128, 3794–3805 (2018).
Hall, C. J. et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ROS production. Cell. Metab. 18, 265–278 (2013).
Shi, H. Z., Wang, D., Sun, X. N. & Sheng, L. MicroRNA-378 acts as a prognosis marker and inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting IRG1. Eur. Rev. Med. Pharmacol. Sci. 22, 3837–3846 (2018).
Pan, J. et al. Immune responsive gene 1, a novel oncogene, increases the growth and tumorigenicity of glioma. Oncol. Rep. 32, 1957–1966 (2014).
Frezza, C. Mitochondrial metabolites: undercover signalling molecules. Interface Focus 7, 20160100 (2017).
Arts, R. J. et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell. Metab. 24, 807–819 (2016).
Tomlinson, I. P. et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410 (2002).
Xiao, M. et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012).
Delage, B. et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 126, 2762–2772 (2010).
Blewett, M. M. et al. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal. 9, rs10 (2016).
Angiari, S. & O’Neill, L. A. Dimethyl fumarate: targeting glycolysis to treat MS. Cell Res. 28, 613–615 (2018).
Kornberg, M. D. et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018).
Brück, J., Dringen, R., Amasuno, A., Pau-Charles, I. & Ghoreschi, K. A review of the mechanisms of action of dimethylfumarate in the treatment of psoriasis. Exp. Dermatol. 27, 611–624 (2018).
Mills, E. A., Ogrodnik, M. A., Plave, A. & Mao-Draayer, Y. Emerging understanding of the mechanism of action for dimethyl fumarate in the treatment of multiple sclerosis. Front. Neurol. 9, 5 (2018).
Zheng, L. et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 6, 6001 (2015).
Sciacovelli, M. & Frezza, C. Oncometabolites: unconventional triggers of oncogenic signalling cascades. Free Radic. Biol. Med. 100, 175–181 (2016).
Rustin, P. et al. Inborn errors of the Krebs cycle: a group of unusual mitochondrial diseases in human. Biochim. Biophys. Acta 1361, 185–197 (1997).
Allegri, G. et al. Fumaric aciduria: an overview and the first Brazilian case report. J. Inherit. Metab. Dis. 33, 411–419 (2010).
Blatnik, M., Thorpe, S. R. & Baynes, J. W. Succination of proteins by fumarate: mechanism of inactivation of glyceraldehyde-3-phosphate dehydrogenase in diabetes. Ann. NY Acad. Sci. 1126, 272–275 (2008).
Adam, J. et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20, 524–537 (2011).
Ooi, A. et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 20, 511–523 (2011).
Sullivan, L. B. et al. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell 51, 236–248 (2013).
Linehan, W. M. et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 374, 135–145 (2016).
Ternette, N. et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 3, 689–700 (2013).
Kerins, M. J. et al. Fumarate mediates a chronic proliferative signal in fumarate hydratase-inactivated cancer cells by increasing transcription and translation of ferritin genes. Mol. Cell Biol. 37, e00079-17 (2017).
Tyrakis, P. A. et al. Fumarate hydratase loss causes combined respiratory chain defects. Cell Rep. 21, 1036–1047 (2017).
Bardella, C. et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 225, 4–11 (2011).
Laukka, T. et al. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 291, 4256–4265 (2016).
Salminen, A., Kauppinen, A. & Kaarniranta, K. 2-Oxoglutarate-dependent dioxygenases are sensors of energy metabolism, oxygen availability, and iron homeostasis: potential role in the regulation of aging process. Cell. Mol. Life Sci. 72, 3897–3914 (2015).
Isaacs, J. S. et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 8, 143–153 (2005).
Yang, M., Soga, T., Pollard, P. J. & Adam, J. The emerging role of fumarate as an oncometabolite. Front. Oncol. 2, 85 (2012).
Sciacovelli, M. et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547 (2016).
Brabletz, T., Kalluri, R., Nieto, M. A. & Weinberg, R. A. EMT in cancer. Nat. Rev. Cancer 18, 128–134 (2018).
Brat, D. J. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498 (2015).
Yogev, O. et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8, e1000328 (2010).
Jiang, Y. et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 17, 1158–1168 (2015).
Sulkowski, P. L. et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 50, 1086–1092 (2018).
Johnson, T. I., Costa, A. S. H., Ferguson, A. N. & Frezza, C. Fumarate hydratase loss promotes mitotic entry in the presence of DNA damage after ionising radiation. Cell Death Dis. 9, 913 (2018).
Ye, D., Guan, K. L. & Xiong, Y. Metabolism, activity, and targeting of d- and l-2-Hydroxyglutarates. Trends Cancer 4, 151–165 (2018).
Chalmers, R. A. et al. d-2-hydroxyglutaric aciduria: case report and biochemical studies. J. Inherit. Metab. Dis. 3, 11–15 (1980).
Duran, M., Kamerling, J. P., Bakker, H. D., van Gennip, A. H. & Wadman, S. K. l-2-Hydroxyglutaric aciduria: an inborn error of metabolism? J. Inherit. Metab. Dis. 3, 109–112 (1980).
Kranendijk, M., Struys, E. A., Salomons, G. S., Van der Knaap, M. S. & Jakobs, C. Progress in understanding 2-hydroxyglutaric acidurias. J. Inherit. Metab. Dis. 35, 571–587 (2012).
Rzem, R. et al. A gene encoding a putative FAD-dependent l-2-hydroxyglutarate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proc. Natl. Acad. Sci. USA 101, 16849–16854 (2004).
Struys, E. A. et al. Mutations in the d-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 76, 358–360 (2005).
Topçu, M. et al. l-2-Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum. Mol. Genet. 13, 2803–2811 (2004).
Kranendijk, M. et al. IDH2 mutations in patients with d-2-hydroxyglutaric aciduria. Science 330, 336 (2010).
Nota, B. et al. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined d-2- and l-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 92, 627–631 (2013).
Dang, L. & Su, S. M. Isocitrate dehydrogenase mutation and R-2-hydroxyglutarate: from basic discovery to therapeutics development. Annu. Rev. Biochem. 86, 305–331 (2017).
Mardis, E. R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
MacIver, N. J., Michalek, R. D. & Rathmell, J. C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 (2013).
Dang, E. V. et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011).
Xu, T. et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 548, 228–233 (2017).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 18, 553–567 (2010).
Losman, J. A. et al. R-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 (2013).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Doedens, A. L. et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat. Immunol. 14, 1173–1182 (2013).
Finlay, D. K. et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453 (2012).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Watanabe, T., Nobusawa, S., Kleihues, P. & Ohgaki, H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 174, 1149–1153 (2009).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Chowdhury, R. et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463 (2011).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 19, 17–30 (2011).
Noushmehr, H. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 17, 510–522 (2010).
Turcan, S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Wang, P. et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32, 3091–3100 (2013).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Zhao, S. et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324, 261–265 (2009).
Intlekofer, A. M. et al. Hypoxia induces production of l-2-hydroxyglutarate. Cell Metab. 22, 304–311 (2015).
Intlekofer, A. M. et al. l-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 13, 494–500 (2017).
Nadtochiy, S. M. et al. Acidic pH Is a metabolic switch for 2-hydroxyglutarate generation and signaling. J. Biol. Chem. 291, 20188–20197 (2016).
Sasaki, M. et al. d-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 26, 2038–2049 (2012).
Tarhonskaya, H. et al. Non-enzymatic chemistry enables 2-hydroxyglutarate-mediated activation of 2-oxoglutarate oxygenases. Nat. Commun. 5, 3423 (2014).
Wang, P. et al. Oncometabolite d-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 13, 2353–2361 (2015).
Inoue, S. et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell 30, 337–348 (2016).
Carbonneau, M. et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat. Commun. 7, 12700 (2016).
Reitman, Z. J. et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA 108, 3270–3275 (2011).
Chesnelong, C. et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro-Oncol. 16, 686–695 (2014).
Grassian, A. R. et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 74, 3317–3331 (2014).
Izquierdo-Garcia, J. L. et al. IDH1 mutation induces reprogramming of pyruvate metabolism. Cancer Res. 75, 2999–3009 (2015).
Chen, J. Y. et al. The oncometabolite R-2-hydroxyglutarate activates NF-κB-dependent tumor-promoting stromal niche for acute myeloid leukemia cells. Sci. Rep. 6, 1 (2016).
Li, F. et al. NADP+–IDH mutations promote hypersuccinylation that impairs mitochondria respiration and induces apoptosis resistance. Mol. Cell 60, 661–675 (2015).
Jiang, B. et al. IDH1 mutation promotes tumorigenesis by inhibiting JNK activation and apoptosis induced by serum starvation. Cell Rep. 19, 389–400 (2017).
Yang, Z. et al. 2-HG inhibits necroptosis by stimulating DNMT1-dependent hypermethylation of the RIP3 promoter. Cell Rep. 19, 1846–1857 (2017).
Kurdistani, S. K. & Grunstein, M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4, 276–284 (2003).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Osinalde, N. et al. Nuclear phosphoproteomic screen uncovers ACLY as mediator of IL-2-induced proliferation of CD4+ T lymphocytes. Mol. Cell. Proteomics 15, 2076–2092 (2016).
Lee, J. V. et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell. Metab. 20, 306–319 (2014).
Zhang, L., Liu, Z., Ma, W. & Wang, B. The landscape of histone acetylation involved in epithelial-mesenchymal transition in lung cancer. J. Can. Res. Ther. 9, S86–S91 (2013).
Mi, W. et al. YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat. Commun. 8, 1088 (2017).
Sivanand, S. et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell 67, 252–265.e56 (2017).
Valls-Lacalle, L. et al. Succinate dehydrogenase inhibition with malonate during reperfusion reduces infarct size by preventing mitochondrial permeability transition. Cardiovasc. Res. 109, 374–384 (2016).
Valls-Lacalle, L. et al. Selective inhibition of succinate dehydrogenase in reperfused myocardium with intracoronary malonate reduces infarct size. Sci. Rep. 8, 2442 (2018).
Kohlhauer, M. et al. Metabolomic profiling in acute ST-segment-elevation myocardial infarction identifies succinate as an early marker of human ischemia-reperfusion injury. J. Am. Heart Assoc. 7, e007546 (2018).
Zhang, J. et al. accumulation of succinate in cardiac ischemia primarily occurs via canonical Krebs cycle activity. Cell Rep. 23, 2617–2628 (2018).
Ariza, A. C., Deen, P. M. & Robben, J. H. The succinate receptor as a novel therapeutic target for oxidative and metabolic stress-related conditions. Front. Endocrinol. (Lausanne) 3, 22 (2012).
Hamel, D. et al. G-protein-coupled receptor 91 and succinate are key contributors in neonatal postcerebral hypoxia-ischemia recovery. Arterioscler. Thromb. Vasc. Biol. 34, 285–293 (2014).
Jean, S. R., Ahmed, M., Lei, E. K., Wisnovsky, S. P. & Kelley, S. O. Peptide-mediated delivery of chemical probes and therapeutics to mitochondria. Acc. Chem. Res. 49, 1893–1902 (2016).
An, J., Rao, A. & Ko, M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 49, e323 (2017).
Vissers, M. C., Kuiper, C. & Dachs, G. U. Regulation of the 2-oxoglutarate-dependent dioxygenases and implications for cancer. Biochem. Soc. Trans. 42, 945–951 (2014).
Zhang, J. et al. Effect of TET inhibitor on bovine parthenogenetic embryo development. PLoS One 12, e0189542 (2017).
Hatch, S. B. et al. Assessing histone demethylase inhibitors in cells: lessons learned. Epigenetics Chromatin 10, 9 (2017).
Maes, T., Carceller, E., Salas, J., Ortega, A. & Buesa, C. Advances in the development of histone lysine demethylase inhibitors. Curr. Opin. Pharmacol. 23, 52–60 (2015).
Pergola, P. E., Spinowitz, B. S., Hartman, C. S., Maroni, B. J. & Haase, V. H. Vadadustat, a novel oral HIF stabilizer, provides effective anemia treatment in nondialysis-dependent chronic kidney disease. Kidney Int. 90, 1115–1122 (2016).
Yeh, T. L. et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem. Sci. 8, 7651–7668 (2017).
Yen, K. et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 7, 478–493 (2017).
Smith, R. A., Hartley, R. C., Cochemé, H. M. & Murphy, M. P. Mitochondrial pharmacology. Trends Pharmacol. Sci. 33, 341–352 (2012).
Smith, R. A., Hartley, R. C. & Murphy, M. P. Mitochondria-targeted small molecule therapeutics and probes. Antioxid. Redox Signal. 15, 3021–3038 (2011).
Yousif, L. F., Stewart, K. M. & Kelley, S. O. Targeting mitochondria with organelle-specific compounds: strategies and applications. Chembiochem 10, 1939–1950 (2009).
Fan, T. W. et al. Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 8, 41 (2009).
Sellers, K. et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 125, 687–698 (2015).
Maher, E. A. et al. Metabolism of [U-13C]glucose in human brain tumors in vivo. NMR Biomed. 25, 1234–1244 (2012).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Joshi, S. et al. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 13, 1895–1908 (2015).
Acknowledgements
This work was supported by a grant to M.P.M. from the Medical Research Council UK (MC_U105663142) and a Wellcome Trust Investigator award to MPM (110159/Z/15/Z). The O’Neill laboratory acknowledges the following grant support: European Research Council (ECFP7-ERC-MICROINNATE), Science Foundation Ireland Investigator Award (SFI 12/IA/1531), GlaxoSmithKline Visiting Scientist Programme and The Wellcome Trust (oneill-wellcometrust-metabolic, grant number 205455).
Author information
Authors and Affiliations
Contributions
All authors wrote, edited and approved final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ryan, D.G., Murphy, M.P., Frezza, C. et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab 1, 16–33 (2019). https://doi.org/10.1038/s42255-018-0014-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-018-0014-7
This article is cited by
-
Microglia and macrophage metabolism: a regulator of cerebral gliomas
Cell & Bioscience (2024)
-
Cancer immunometabolism: advent, challenges, and perspective
Molecular Cancer (2024)
-
Cancer cell metabolism and antitumour immunity
Nature Reviews Immunology (2024)
-
Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners
Nature Communications (2024)
-
A break in mitochondrial endosymbiosis as a basis for inflammatory diseases
Nature (2024)