Abstract
Escherichia coli glutamate decarboxylase (EcGadB), a pyridoxal 5’-phosphate (PLP)-dependent enzyme, is highly specific for L-glutamate and was demonstrated to be effectively immobilised for the production of γ-aminobutyric acid (GABA), its decarboxylation product. Herein we show that EcGadB quantitatively decarboxylates the L-isomer of D,L-2-amino-4-(hydroxyphosphinyl)butyric acid (D,L-Glu-γ-PH), a phosphinic analogue of glutamate containing C-P-H bonds. This yields 3-aminopropylphosphinic acid (GABA-PH), a known GABAB receptor agonist and provides previously unknown D-Glu-γ-PH, allowing us to demonstrate that L-Glu-γ-PH, but not D-Glu-γ-PH, is responsible for D,L-Glu-γ-PH antibacterial activity. Furthermore, using GABase, a preparation of GABA-transaminase and succinic semialdehyde dehydrogenase, we show that GABA-PH is converted to 3-(hydroxyphosphinyl)propionic acid (Succinate-PH). Hence, PLP-dependent and NADP+-dependent enzymes are herein shown to recognise and metabolise phosphinic compounds, leaving unaffected the P-H bond. We therefore suggest that the phosphinic group is a bioisostere of the carboxyl group and the metabolic transformations of phosphinic compounds may offer a ground for prodrug design.
Similar content being viewed by others
Introduction
The pyridoxal 5′-phosphate (PLP)-dependent enzyme glutamate decarboxylase (L-glutamate 1-carboxy-lyase; Gad; E.C. 4.1.1.15) catalyses the irreversible decarboxylation on C-α of the proteinogenic amino acid L-glutamate to yield γ-aminobutyrate (GABA). The enzyme from Escherichia coli is active only on the L-isomer of glutamate, with the D-isomer neither being a substrate nor an inhibitor1,2. The E. coli enzyme, intensively studied at the biochemical and structural level, is a homohexamer, active in the pH range 3.8–5.4, positively responding to chloride ions, and undergoing a remarkable auto-inactivation above pH 5.53,4. Lammens et al.5 were the first to show that E. coli GadB (EcGadB) can be efficiently immobilised and used as an excellent tool for the bio-based synthesis of GABA. This study was followed by several others6,7,8,9. The interest residing in the fact that L-glutamate, a non-essential amino acid and one of the most abundant in proteins, can be extracted from waste biomass and as an alternative source for the production of a valuable compound, such as GABA, from which 2-pyrrolidone, N-methyl-pyrrolidone, N-vinyl-pyrrolidone as well as polymers and pharmaceuticals (e.g., cotinine, piracetam, iodopovidone) can be derived10,11. In addition, EcGadB was engineered to optimise GABA synthesis for large scale production in particular by rendering the enzyme less prone to rapidly auto-inactivate at pH > 5.54,11,12.
Even though EcGadB decarboxylates at a significant rate only L-glutamate, its physiological substrate (Km ~ 1 mM)1,13, it was reported that the enzyme slowly decarboxylates glutamate analogues such as: L-glutamate-γ-methylester, threo-β-hydroxy-glutamate, 3-fluoro-L-glutamate, γ-methylene-glutamate, L-homocysteine sulphinic acid and L-homocysteic acid1,14,15,16,17. Except for the latter two, none of the listed compounds is a naturally occurring amino acid.
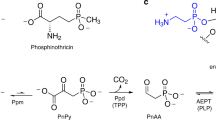
Naturally occurring amino acids also include some phosphonic and phosphinic compounds, i.e., containing a carbon–phosphorous bond (C–P). Phosphonates can mimic a phosphate monoester or a tetrahedral intermediate (or reaction transition states) of the carboxyl group transformations18,19,20. These unusual phosphorus-containing compounds, naturally produced by many micro-organisms, are synthesised chemically in the pharmaceutical and agro-chemical industry and have found application in medicine and agriculture21,22. Notable examples include the antibiotic fosfomycin, the antimalarial fosmidomycin and the herbicides bialaphos (PTT; Fig. 1) and phosalacine, which are tripeptides containing phosphinothricin (PT; Fig. 1) followed by alanyl-alanine and alanyl-leucine residues, respectively. PT is a glutamate analogue known to inhibit glutamine synthetase by mimicking the phosphorylated intermediate of glutamate formed during the enzymatic reaction23,24. Bialaphos was first shown to be produced by Streptomyces viridochromogenes and Streptomyces hygroscopicus25,26. In both micro-organism the biosynthetic pathway leading to bialaphos was later characterised27. Along this pathway, L-2-amino-4-(hydroxyphosphinyl)butyric acid (also known as desmethylphosphinothricin, DMPT; Fig. 1), hereafter L-Glu-γ-PH, was shown to be a key intermediate, becoming detectable only in some mutant strains28.
The herbicide bialaphos (PTT), produced by S. viridochromogenes and S. hygroscopicus, is a tripeptide containing L-phosphinothricin (PT) followed by alanyl-alanine (L-Ala-LAla) residues. In the biosynthetic pathway leading to PTT, desmethylphosphinothricin (DMPT; L-Glu-γ-PH) is a key intermediate, from which 3-aminopropylphosphinic acid (3-APPA; GABA-PH) and Succinate-PH can be derived following in order DMPT decarboxylation and oxidation, respectively.
L-Glu-γ-PH is a phosphinic analogue of glutamate and to date very little is known about its biological activity: when tested for its in vitro ability to bind group III metabotropic L-Glu receptors, which are mostly presynaptic and controlling glutamate release at the synaptic level, L-Glu-γ-PH potency was found in the same range as that of glutamate29. Recent work carried out in our laboratories suggests that L-Glu-γ-PH has antibacterial activity30. Except for these reports, the substrate/inhibitor properties of L-Glu-γ-PH on glutamate metabolising enzymes, including bacterial glutamate decarboxylase, have never been studied.
In the present work, we investigate the substrate properties of L-Glu-γ-PH on EcGadB and, by exploiting the specificity of the enzyme for the L-isomer, develop a bio-based route for the kinetic resolution of the D-isomer from D,L-2-amino-4-(hydroxyphosphinyl)butyric acid (D,L-Glu-γ-PH). The efficient enzymatic decarboxylation of L-Glu-γ-PH yields 3-aminopropylphosphinic acid (Fig. 1), hereafter GABA-PH, an agonist of GABAB receptor shown to be more potent than Baclofen in vitro31. Moreover, we demonstrate that GABA-PH can be further metabolised to yield the phosphinic analogue of succinate (Succinate-PH; Fig. 1).
Results
Retrosynthetic analysis of D-Glu-γ-PH
The earlier unknown D-Glu-γ-PH (1) may be prepared chemically (Fig. 2) and key precursor is methyl N-Cbz-D-vinylglycine (2), which can be synthesised following the protocols developed and used for the corresponding L-isomer32. To prepare methyl N-Cbz-L-vinylglycine different starting compounds, including amino acids, were used and reported elsewhere32 and many of these synthetic approaches may be directly applied to prepare (2). In brief, the most well-known pathways are shown as B, C, D in Fig. 2. Pathway (D), starting from D-Met, follows a classical and widely used 3-step Rapoport synthesis32. Alternatively, (2) can be prepared from commercially available D-3-aminobutyrolactone hydrochloride (3) in six steps, either following pathway (C) via selenide (4)33 or via sulfoxide (5), following pathway (B), which also consists of six steps34. Besides, pathway (C) requires working with diphenyldiselenide, which is toxic for vertebrates and environment33.
(A) From racemic D,L-Glu-γ-PH (this work, in blue) in one enzymatic step; (B) from D-2-aminobutyrolactone hydrochloride (3) via o-nitrophenyl-sulfoxide (5) in six steps; (C) from D-2-aminobutyrolactone hydrochloride (3) via phenylselenide (4) in six steps; (D) from D-methionine via sulfoxide (6) in three steps. Pathways (B)–(D) lead to the same key precursor methyl N-Cbz-D-vinylglycine (2).
Conversion of (2) into target D-Glu-γ-PH (1) consists in anti-Markovnikov radical addition of H3PO2 to the double bond of (2) and subsequent removal of the protecting groups, as described for L-Glu-γ-PH in Supplementary Methods and elsewhere29. Hence, chemical syntheses of D-Glu-γ-PH, are laborious, consist of five to eight steps and can provide target D-Glu-γ-PH at a rather low overall yield.
The stereoselectivity of EcGadB could alternatively be exploited for the kinetic resolution of D,L-Glu-γ-PH using EcGadB (Fig. 2, pathway A) as a fast, quantitative and inexpensive bio-based approach to synthesise unknown D-Glu-γ-PH and, at the same time, to obtain GABA-PH, the anticipated decarboxylation product of L-Glu-γ-PH by EcGadB. This working hypothesis was based on the assumption that the single-charged phosphinic group mimics the flat carboxyl group not only in the global charge (i.e., single), but also because of its flattened tetrahedral conformation (Supplementary Fig. 1) as suggested by the crystallographic evidence on the bond lengths in the β-phosphinic analogue of aspartate35.
Given that D,L-Glu-γ-PH is readily available with high yield via four steps36 or via a three-step one-pot synthesis37 from dibutyl ester of vinylphosphonic acid, the bioconversion protocol, herein used, is proposed to leave D-Glu-γ-PH unmodified from D,L-Glu-γ-PH, assuming that the L-isomer is quantitatively converted into GABA-PH by EcGadB. The downstream purification from the reaction mixture would therefore allow to prepare unknown D-Glu-γ-PH and commercially unavailable, biologically active GABA-PH.
Preliminary studies on L-Glu-γ-PH as substrate of EcGadB
Substrate properties of L-Glu-γ-PH (DMPT; Fig. 1) in EcGadB reaction were not self-evident, since phosphinothricin (PT; Fig. 1) and the γ-phosphonic counterpart of L-Glu-γ-PH, i.e. 2-amino-4-phosphonobutyric acid, were shown to be neither substrates nor inhibitors of the enzyme38,39. Initially, we investigated if EcGadB can bind D,L-Glu-γ-PH and specifically decarboxylate it into GABA-PH by assessing the course of the reaction using thin layer chromatography (TLC). These experiments demonstrated the formation of a compound having the Rf value similar to that of GABA (Supplementary Fig. 2). To confirm that the newly formed compound is GABA-PH, we performed 1H NMR analysis of EcGadB reaction mixture with D,L-Glu-γ-PH as a substrate in Pyridine/HCl buffer. Glu-γ-PH and GABA-PH have characteristic signals that only partly overlap in 1H NMR spectra (Supplementary Fig. 3) that clearly showed that the incubation of D,L-Glu-γ-PH with EcGadB results in the formation of GABA-PH (Supplementary Fig. 3). In addition, TLC provided additional evidence that the γ-phosphonic counterpart of L-Glu-γ-PH is not a substrate of EcGadB, as already reported38,39.
Modelling of L-Glu-γ-PH binding in EcGadB active site
To compare the binding mode and interactions of L-Glu and L-Glu-γ-PH to EcGadB active site, molecular dynamics (MD) simulations40 were carried out by setting a constant pH of 4.5 and using the Protein Data Bank code 1PMM41. The external aldimine adducts PLP-L-Glu and PLP-L-Glu-γ-PH were drawn using the coordinates of the co-crystallised PLP and acetic acid (ACY) in 1PMM41. The trajectory analyses for the L-Glu with its cognate protein showed a good stability with a root mean square fluctuation (RMSF) of 1.12, 0.46 and 0.76 Å for the enzyme, the PLP-Lys276 internal aldimine and the PLP-L-Glu external aldimine adduct, respectively. The inspection of the binding modes highlighted a series of key contacts that are characteristic of the PLP-L-Glu adduct into the binding site: the PLP moiety, which was superimposable with the binding conformation reported in 1PMM, is stabilised by hydrophobic contact with Gln163 and Ala245 side chains. Furthermore, the PLP hydroxyl group is engaged in intramolecular H-bond with the aldimine nitrogen atom. The phosphate group is involved in H-bonds network: the OP1 with Ser127 and Ser128, the OP2 with Ser273* and His275, the OP3 with Ser318*. The α-carboxyl group is perpendicular to the PLP plane and too far (4.6 Å) from Gln163 to establish a polar contact. The γ-carboxylic moiety is engaged in a network of H-bonds involving Asp86*, Phe63 and Cys64, in good accordance with the literature data (Fig. 3, left panel)41.
Snapshots of the molecular dynamic simulations. a Proposed binding mode for PLP-L-Glu adduct (cyan). b Proposed binding mode for PLP-L-Glu-γ-PH adduct (cyan). Residues involved in interaction are reported as stick, yellow for chain B and green for chain A of the functional dimer41. PLP and acetic acid (ACY) of 1PMM are also reported as magenta lines. The enzyme is depicted as grey cartoon and the H-bonds are reported as yellow dotted lines. Active site residues contributed by the other subunit in the functional dimer are also labelled with an asterisk.
The same analyses were performed for L-Glu-γ-PH. The RMSF values were 1.35, 0.47 and 1.07 Å for the enzyme, the PLP-Lys276 internal aldimine and the PLP-L-Glu-γ-PH external aldimine, respectively. The contacts that stabilised the PLP-L-Glu-γ-PH adduct (Fig. 3, right panel) resembled the interactions already reported for the PLP-L-Glu (Fig. 3, left panel). Also, the polar contacts that stabilised the phosphinic moiety of L-Glu-γ-PH were similar, though not identical, to those described for the L-Glu γ-carboxylic acid and involving Thr62, Phe63, Cys64 and Asp86*. It was in addition noticed that during the simulations the distances between the Lys276 ε-amino group and the external aldimine C-α atom of the amino acid substrate were always close to 3.5 Å, in agreement with the catalytic role of this residue42. Together with the preliminary evidence (Supplementary Figs. 2 and 3), the good superimposition between the L-Glu and L-Glu-γ-PH binding modes (Fig. 3) strengthens the working hypothesis that the L-Glu-γ-PH is an EcGadB substrate. However, some noticed differences may account for the observed changes in the binding affinity (next section). Noteworthy, the same sort of analysis failed to provide a reliable model for the binding of the γ-phosphonic counterpart of L-Glu-γ-PH to the EcGadB active site. Thus supporting previous studies36,37 and the TLC analysis (Supplementary Fig. 2).
L-Glu-γ-PH is a substrate of EcGadB and GABA-PH of GABase
L-Glu-γ-PH is not commercially available. In this work it was synthesised according to a published protocol29, with some modifications (Supplementary Methods), and, supported by the preliminary data described in the previous section, its substrate properties on EcGadB were studied.
In order to obtain the kinetic parameters of the EcGadB reaction with L-Glu-γ-PH, we initially investigated whether the GABase assay, routinely used to measure GABA formed following the decarboxylation of L-Glu by bacterial EcGadB activity13,42,43, could be employed also for detecting the products of the reaction with L-Glu-γ-PH. GABase is a commercial crude preparation of PLP-dependent GABA-transaminase and NADP+-dependent succinic semialdehyde dehydrogenase from Pseudomonas fluorescens: it works in the neutral-to-mild alkaline pH range by converting GABA into succinic semialdehyde (via GABA-transaminase activity)43,44 which is then irreversibly oxidised under the assay conditions into succinic acid (via succinic semialdehyde dehydrogenase) with quantitative formation of NADPH. The latter reduced cofactor can be easily detected spectrophotometrically13,45,46. It was therefore of interest to find out whether GABA-PH, generated by the irreversible decarboxylation of L-Glu-γ-PH, could be detected through GABase. Succinate semialdehyde-PH (SSA-PH), the phosphinic analogue of succinic semialdehyde and transamination product of GABA-PH, has in its structure a phosphinic moiety with a P–H bond and a carbonyl group, both possessing a high reducing potential. In order to assess to which one of the two groups (phosphinic or carbonyl) the formation of NADPH could be assigned, 31P NMR spectroscopy analysis was performed before and after GABase treatment of the EcGadB reaction mixture (Fig. 4) as follows: (1) the conversion of D,L-Glu-γ-PH (spectrum a in Fig. 4) into GABA-PH (spectrum b in Fig. 4) was allowed to occur in the presence of EcGadB, at pH 4.6, and spectrum b was run using half of the reaction mixture, after removing EcGadB by ultrafiltration; (2) the remainder of the reaction mixture was dried under vacuum and resuspended in an isovolume of GABase assays solution to allow GABA-PH to be converted into Succinate-PH by GABase46.
The EcGadB (0.09 mg ml−1) decarboxylation reaction of D,L-Glu-γ- PH (18 mM) was carried out at pH 4.6, in 10 mM sodium acetate buffer (pH adjusted with small additions of diluted HCl) and then halted after 4 h by removing EcGadB by ultrafiltration. Half of the flow-through was dried and resuspended in an isovolume of GABase assay solution where the two indicated enzymatic reactions were run one-pot, with succinic semialdehyde dehydrogenase activity making the whole process irreversible. 31P NMR spectra of: (a) EcGadB reaction mixture at time zero; (b) EcGadB reaction mixture after interruption by enzyme removal; (c) reaction mixture after GABase treatment; (d) model mixture of L-Glu-γ-PH, GABA-PH, and Succinate-PH.
For comparative purposes, the latter reaction was terminated before completion to still allow detection of the GABA-PH signal in the final 31P NMR spectrum. Before running the latter 31P NMR spectroscopy analysis, GABase was removed by ultrafiltration. The signals of the three compounds present after the GABase reaction are in the region typical for the expected phosphinic compounds (spectrum c in Fig. 4) and the identity of each was further corroborated by 31P NMR spectroscopy analysis of a mixture of model compounds (spectrum d in Fig. 4).
Thus, the 31P NMR data provided incontrovertible evidence that the formation of NADPH, as detected spectrophotometrically (data not shown), was due to the oxidation of the carbonyl moiety of 3-(hydroxyphosphinyl)propanal (SSA-PH in Fig. 4) yielding Succinate-PH. To the best of our knowledge, while previously hypothesised47, this is the first report where direct evidence of a selective NAD(P)+-dependent enzymatic oxidation of a functional group in the side chain of a phosphinic acid, which leaves unaffected the P–H bond, is provided.
The substrate properties of L-Glu-γ-PH for EcGadB were therefore assessed by kinetic analysis, using GABase which allowed to quantitatively measure the GABA-PH produced by detecting spectrophotometrically the NADPH produced. These results were compared with those obtained with L-Glu, the physiological substrate of EcGadB (Table 1).
Based on the results shown in Table 1, it turned out that EcGadB is >200 folds less efficient in decarboxylating L-Glu-γ-PH if compared with L-Glu. This difference could be primarily assigned to the increase in Km (Table 1) and explained, at least in part, by some differences in the anchoring of the distal carboxyl and phosphinic groups in EcGadB active centre, as suggested by the modelling studies (Fig. 3).
Enantioselective decarboxylation of D,L-Glu-γ-PH by EcGadB
In the previous sections, evidence has been provided that EcGadB can accept L-Glu-γ-PH as a substrate and decarboxylate it into GABA-PH. At the same time, pathway A in Fig. 2 shows that D-Glu-γ-PH could be easily prepared starting from D,L-Glu-γ-PH, by taking advantage of the well-known selectivity of the enzyme for the L-isomer of glutamate1,2, which would therefore yield GABA-PH and leave unchanged D-Glu-γ-PH.
In order to assess the feasibility of this approach, a preliminary experiment (on a small scale; 100 µL) using EcGadB and D,L-Glu-γ-PH (0.4 mol L−1) as substrate was carried out for 2 h, under the conditions described in the “Methods” section. The production of GABA-PH followed hyperbolic kinetics and the amount present at 2 h (0.197 mol L−1) was almost quantitative, according to the GABase assay, with respect to the amount (0.200 mol L−1) of L-isomer present in the original reaction mixture. The kinetics of GABA-PH formation using GABase (Supplementary Fig. 4) indicated that close to 50% of D,L-Glu-γ-PH remained unchanged. This provided a strong indication that indeed EcGadB could decarboxylate only the L-isomer of Glu-γ-PH and therefore be exploited to obtain preparative amounts of D-Glu-γ-PH and GABA-PH. The key parameter of this enzymatic reaction, as already reported for the preparative synthesis of GABA from L-Glu5,12, was pH, which was controlled at time intervals in order to be within the range 4.6–5.0, where EcGadB displays maximal activity12,13.
Preparative decarboxylation was therefore set up by scaling up the above reaction 25 times (final volume 2.5 mL). In this experiment 31P NMR spectroscopy was employed in a real-time mode to allow a direct and immediate detection of GABA-PH formation from D,L-Glu-γ-PH. As expected, the amount of Glu-γ-PH in the reaction mixture decreased with time and a concomitant increase of GABA-PH was observed, which after 3 h reached 93% of the theoretical value (Fig. 5).
The kinetics of the decarboxylation of D,L-Glu-γ-PH by EcGadB yielding GABA-PH are shown. The time point (in minutes) at which each measurement was done is shown on the right and next to it (in brackets) is provided the Glu-γ-PH/GABA-PH ratio obtained by peak integration. Fragments of the 31P NMR spectra are shown as a stack.
The reaction was allowed to proceed for additional 2 h during which the residual L-Glu-γ-PH was decarboxylated and the amount of GABA-PH reached 98.2% of the theoretical value.
The preparative experiment was performed in triplicate, with kinetics and final yields closely resembling those shown in Fig. 5.
These results provided a strong indication that indeed half of D,L-Glu-γ-PH was not decarboxylated (very likely the D-isomer) and that the bioconversion could be employed to obtain preparative amounts of GABA-PH and D-Glu-γ-PH, as confirmed by physical-chemical techniques and enzymatic assays (next section).
Isolation of enzymatically obtained D-Glu-γ-PH and GABA-PH
After the removal of EcGadB through ultrafiltration, GABA-PH and D-Glu-γ-PH (according to the enantioselectivity of the enzyme) were isolated from the reaction mixture using ion-exchange chromatography on Dowex 50WX8 (H+-form). Elution with water provided crude D-Glu-γ-PH which was contaminated with PLP, originally present in the reaction mixture (see “Methods” section). GABA-PH hydrochloride was recovered with 0.5 N HCl. It was then converted to zwitterion and recrystallised. This sample was identical to chemically synthesised GABA-PH as assessed by 1H, 31P and 13C NMR spectra (Supplementary Fig. 5), TLC and melting point (see “Methods” section).
In order to separate D-Glu-γ-PH and PLP, the above crude sample was treated with an excess of aminooxyethyl putrescine (AOEPUT) as depicted in Supplementary Fig. 6. AOEPUT does not react with Glu-γ-PH, but forms oxime with PLP fast and quantitatively. The two amino groups of both free AOEPUT and AOEPUT-PLP oxime ensure tight binding to Dowex 50WX8, making it impossible to elute them with water, but on the contrary allowed to recover D-Glu-γ-PH from the resin. After crystallisation, D-Glu-γ-PH was analysed by 1H, 31P and 13C NMR spectroscopy (Supplementary Fig. 7) and displayed [α]D20 value of −20.4°, (c 1.0, H2O). The direction of the rotation clearly indicated it to be the D-isomer (see “Methods” section) because its value was in full agreement with the rotation [α]D20 value of +20.9°, (c 1.0, H2O) of L-Glu-γ-PH chemically synthesised in this work (Supplementary Methods). Notably, the latter value is rather different from that reported in the literature for L-Glu-γ-PH [α]D20 +7.9° (c 0.4, H2O)29. Thus the herein enzymatically prepared D-Glu-γ-PH has the same specific rotation value of the chemically synthesised L-Glu-γ-PH, though in the opposite direction. This latter observation provides an unquestionable indication that the two isomers prepared in this work are indeed the exact compounds. To further prove this, the reactivity of D-Glu-γ-PH towards EcGadB was studied. Any attempt to detect GABA-PH from D-Glu-γ-PH failed, even when the reaction was allowed to occur overnight, in line with the well-known stereospecificity of the enzyme (data not shown).
Antimicrobial activity of Glu-γ-PH enantiomers
To assess the contribution of each isomer to the antimicrobial activity of D,L-Glu-γ-PH30, the minimum inhibitory concentration (MIC) of the racemate as well as that of the L- and D-isomers were assayed on the E. coli K12 strain MG1655 (Table 2).
The results in Table 2 provided preliminary, but clear indication that the L-isomer was the only molecule responsible for the antibacterial activity and that its MIC is in the same range of a well-known antibiotic, i.e., ampicillin. Notably, to reach MIC90, the concentration of D,L-Glu-γ-PH, had to be twice as high as that of L-Glu-γ-PH, thus ensuring that the L-isomer was at the effective working concentration. The MIC value of the racemate is thus compatible with one isomer (L) being active and the other one (D) being inactive. The interval in the MIC90 of the racemate suggests that the D-isomer may slightly interfere with the L-isomer transport, a finding non uncommon in transport studies48. On the other hand, the D-isomer, when assayed alone, exhibited a hardly detectable activity, i.e., it caused only 4% of inhibition of growth at the concentration of 128 µg mL−1. Thus, the MIC analysis confirmed that via kinetic resolution the distomer, i.e. D-Glu-γ-PH, could be obtained through a bioconversion without the need of chemically synthesising it and it was devoid of a significant antibacterial activity.
Discussion
Through a combination of enzymology, NMR spectroscopy, molecular modelling and microbiological assays, we provide evidence that in some phosphinic compounds, i.e. containing C–P–H bonds, the phosphinic moiety acts as a bioisostere of the carboxyl group, making them substrates of the relevant enzymes. In particular, as part of the kinetic resolution of D,L-Glu-γ-PH using EcGadB, it is possible to prepare unknown D-Glu-γ-PH and biologically active GABA-PH and offer an excellent route to their synthesis.
Our working hypothesis was based on the assumption that the single-charged phosphinic group mimics the flat carboxyl group not only in the global charge (i.e., single) but also because of its flattened tetrahedral conformation (Supplementary Fig. 1). On the contrary, the double-charged tetrahedral phosphonic group turns out to be a bad mimetic of the carboxyl group (Supplementary Fig. 1). Our hypothesis was also in good agreement with previous reports showing that (1) the γ-phosphonic analogue of L-Glu, as well as PT, is neither substrates, nor inhibitor of GadB38,39; (2) α-phosphinic analogues of amino acids, but not α-phosphonic analogues, are substrates of PLP-dependent alanine transaminase, methionine γ-lyase and tyrosine phenol-lyase47,49,50; (3) that group III metabotropic L-Glu receptors display an affinity towards L-Glu-γ-PH comparable to that of L-Glu29 and (4) D,L-Glu-γ-PH can act as a weak amino donor to the α-keto precursor of PT in E. coli GABA-transaminase51.
Starting D,L-Glu-γ-PH is readily available with high yield36,37, and the downstream purification from the bioconversion reaction mixture provides D-Glu-γ-PH with a preparative yield of 73%. Besides, as part of the kinetic resolution, GABA-PH, an agonist of GABAB receptor shown to be more potent than Baclofen in vitro31, is formed. In analogy with previously published work on the bioconversion of L-Glu with immobilised EcGadB5, preliminary data in solution indicate that the bioconversion reaction of D,L-Glu-γ-PH is feasible also in water (D. De Biase, personal communication), by controlling the pH of the reaction with a pH-stat.
Taking into account that L-Glu-γ-PH and some of its derivatives are currently under investigation as antibacterials30, it was of key importance to assess that the D-Glu-γ-PH, the distomer, was neither active nor displayed opposite or significantly negative effects in the in vivo testing. Moreover, the scalable bioconversion protocol described in this work makes D-Glu-γ-PH easily accessible to study its properties in relation to other currently unforeseen applications.
It is also important to recall that the substrate properties of the phosphinic analogue of succinic semialdehyde (SSA-PH) as occurring in the GABase reaction were not self-evident, since it is known that phosphorous acid (H3PO3), having P–H bond, is efficiently oxidised to phosphoric acid (H3PO4) by NAD+-dependent dehydrogenase encoded by the ptxD gene52. Therefore, our results with P. fluorescens GABase confirm that in addition to the PLP-dependent enzyme (GABA-transaminase), a NADP+-dependent dehydrogenase (Succinic semialdehyde dehydrogenase) can accept a phosphinic compound as substrate. The latter represents the first example of the NADP+-dependent enzymatic transformation in the side chain of alkylphosphinic acids having the P–H bond with high reducing potential.
Our findings, summarised in Fig. 6, suggest that enzymatic approaches to the synthesis of phosphinic compounds are feasible.
EcGadB converts at mildly acidic pH (yellow background) the L-isomer of D,L-Glu-γ-PH (carbon skeleton in yellow on the outmost right), quantitatively yielding GABA-PH (carbon skeleton in yellow on the bottom left) and leaving D-Glu-γ-PH unmodified (carbon skeleton in cyan on the outmost right). The former becomes a substrate of GABase that yield 3-(hydroxyphosphinyl)propionic acid (i.e., Succinate-PH shown with carbon skeleton in yellow on the top centre) in a coupled reaction that occurs at mildly alkaline pH (blue background).
Our data suggest that phosphinic compounds can be metabolised intracellularly to such a level that yields metabolites which eventually target enzymes different from the initially processed aminophosphinate; this may turn out to be a productive strategy in prodrug design. To date, the only known example of this process is the intracellular transformation of the α-phosphinic derivative of L-Ala (Ala-PH) into the phosphinic analogue of pyruvate, a well-characterised nM inhibitor of pyruvate dehydrogenase53, with antitumor potential20. Thus, respect to compounds containing bonds C–P–C (i.e., methylphosphinic) or C–P–O (phosphonic), thoroughly investigated as such and as a key element of peptidomimetics19, the biochemical and medicinal potential of compounds having C–P–H (phosphinic) bonds has received very little attention.
Methods
Materials
D,L-Glu-γ-PH was synthesised as described earlier37. Synthesis of L-Glu-γ-PH was performed following a published protocol29, with some modifications, as described in Supplementary Information. 3-(Hydroxyphosphinyl)propionic acid (Succinate-PH) was synthesised as described elsewhere54 and the product was purified from contaminating H3PO2 by column chromatography on Kieselgel 60 (40–63 μm, Merck, Germany), using CHCl3:MeOH:AcOH, 7:2:1, v/v for elution. N-(2-aminooxyethyl)-putrescine trihydrochloride (AOEPUT) was synthesised as previously described55. Ion-exchange chromatography was carried out on Dowex 50WX8 (H+-form, 100–200 mesh; BioRad, USA). Elution conditions are specified in the text. TLC was carried out on Kieselgel 60 F254, or Cellulose F254 plates (Merck, Germany) in i-PrOH:25%NH4OH:H2O, 7:1:2, v/v. Amino acids were detected on TLC plates following ninhydrin staining (0.4% in acetone).
For the expression, purification and assay of EcGadB, ingredients and chemicals were from the following suppliers: the ingredients for bacterial growth were from BD-Difco (Becton, Dickinson and Company); streptomycin sulfate was from US Biochemical Corp. (Cleveland, OH, USA); ampicillin was from Roche; ion-exchange chromatography was performed using pre-packed HiPrep DEAE FF 16/10 (20 mL) from GE Healthcare Life Sciences; pyridine was from Carlo Erba; Vitamin B6, potassium dihydrogen phosphate, dipotassium hydrogen phosphate and kanamycin were from Fluka; sodium glutamate, sodium acetate, acetic acid, sodium chloride, GABA and PLP were purchased either from Merck or from Sigma-Aldrich. GABase was from Sigma-Aldrich. Vivaspin (4 mL and 500 µL; 30 and 10 kDa cutoff, respectively) ultrafiltration devices were from Sartorius Stedim Lab Ltd, UK.
For antimicrobial testing, Minimal Medium E56, supplemented with 0.4% glucose, was used. Glucose was from Carlo Erba. All other ingredients were either from VWR or Applichem. The E. coli strain tested was the K12 strain MG165557,58.
Analytical techniques
1H, 13C and 31P NMR spectra of D-Glu-γ-PH and GABA-PH were recorded on a Bruker 300 AM-300, while for NMR analysis of enzymatic reactions a Bruker 500 UltraShieldTM was used. Spectra were measured in D2O, or in H2O/D2O mixture, by using with sodium 3-trimethyl-1-propanesulfonate as internal standard, or 85 % H3PO4 as an external standard. Chemical shifts are given in ppm. Optical rotations were recorded on a 341 Polarimeter (Perkin-Elmer); solvents and concentrations are indicated in the text. Melting points were determined in open capillary tubes on Electrothermals Mel-Temp 1202D instrument and are uncorrected.
Enzymatic assays and calculation of kinetic parameters
To find out the reaction conditions for the preparative scale experiments, the kcat and Km values for L-Glu were obtained by following EcGadB (0.55 μg) reaction in the presence of L-Glu (from 0.156 to 20 mM) in a final reaction volume of 250 µL of 0.1 M Pyridine/HCl, pH 4.6 containing 0.1 mM DTT and 40 µM PLP. The experiment was performed in duplicate. The kcat and Km values for L-Glu-γ-PH were then determined in the same buffer system. In the latter case, GadB (13 μg) was incubated with L-Glu-γ-PH (from 6 to 220 mM) in a final reaction volume of 160 µL. The experiment was performed in duplicate. For both substrates, the rates were determined by halting the reaction, which consisted in transferring aliquots (50 or 25 μL) of the reaction mixture into four volumes (200 or 100 μL, respectively) of 0.1 M HEPPS buffer, pH 8.6, immediately followed by vigorous vortexing. At each tested concentration, three time points (for L-Glu 1–2–3 min; for L-Glu-γ-PH 3–7–10 min) were taken.
GABA (from L-Glu) or GABA-PH (from L-Glu-γ-PH) content was assayed with the GABase assay by transferring 10 or 50 μL of each halted reaction into 100 μL of GABase solution containing 1.25 µL of activated GABase (0.02 Units µL−1). The incubation was carried out for 60 min at 37 °C to allow the full transamination and oxidation by GABase of GABA (or GABA-PH) into succinate (or Succinate-PH) and the concomitant formation of NADPH. The latter was then quantified by directly measuring its 340 nm absorbance by applying the molar absorption coefficient (ε) NADPH340 = 6220 M−1 cm−1.
The readings of the NADPH produced at each time point, for each substrate concentration, were fitted by linear regression and converted back into nmoles of GABA (or GABA-PH) s−1. The reaction rates at increasing substrate concentration were then fitted to the standard Michaelis–Menten equation as in GraphPad Prism 4.0. (GraphPad Software, San Diego, CA).
Determination of minimum inhibitory concentration
The MIC of D,L-Glu-γ-PH, L-Glu-γ-PH and D-Glu-γ-PH that inhibits the growth of the E. coli K12 strain MG1655 was assessed using the broth dilution method59 with some minor modifications, i.e., MIC90 was calculated in this work, i.e., meaning that the indicated value is the concentration that inhibits 90% of the growth (as assessed at O.D.600) at a specific time from the inoculum (22 h). MIC90 is derived by the following equation: % inhibition = [1 − (OD600treated/OD600untreated)] × 100.
Briefly, overnight cultures (2 mL) of E. coli K12 strain MG1655 were centrifuged at 3500 rpm for 15 min at 15 °C and the cell pellets were resuspended in an equivalent volume of physiological solution (0.9% NaCl) and the O.D.600 was brought to 1.0. The resuspended cells were inoculated (1:25) in 2 mL of Minimal EG pH 7 and grown at 37 °C under moderate shaking up to O.D.600 = 0.5 (7–8 h from the inoculum) and then diluted (1:25) in 3 mL of Minimal medium EG pH 756 and inoculated (1:10) in a 96-well microplate (FalconTM) previously set up with the appropriate serial dilutions of the compound(s) to be tested. The final volume in each well was 200 µL. The number of colony forming units/ml at time zero was between 0.5 and 1 × 106, following the CLSI guidelines for antimicrobial susceptibility testing60. For each compound tested, 2–4 independents assays were performed.
The microplate was incubated at 37 °C for 24 h and reading carried out on microplate readers (Sunrise, Tecan or Varioskan Lux, Thermo Scientific) and the OD600 was automatically recorded every hour. Before each reading, the microplate was set to shake 10 s in order to resuspended the bacteria.
Kinetic resolution of D,L-Glu-γ-PH by EcGadB
The reaction mixture (100 µL for analytical purposes or 2.5 mL for preparative purposes) consisted of 200 mM Pyridine/HCl buffer, pH 4.6, containing 1 mM PLP, 0.1 mM DTT, 0.4 M of D,L-Glu-γ-PH to which GadB was added to a final concentration of 1 µg µL−1.
The analytical reactions (100 µL) were performed at 37 °C and at time intervals (0, 2, 5, 10, 20, 30, 60, 120 min). During the reaction, the pH was periodically adjusted to the value 4.6–5.0 by stepwise addition of 1 N HCl (total: 10 µL). At the indicated times, aliquots (5 μL) were transferred in 50 µL of 10 mM NaOH and then 2 µL analysed for GABA content with the GABase assay (see previous section). The experiment was performed in duplicate.
The preparative reaction was performed at 37 °C, adjusting the pH periodically to the value of 4.6–5.4 by adding 1 N HCl (total: 80 μL). At time intervals (0, 2, 5, 10, 20, 40, 60, 90, 120, 150, 180, 240, 300 min), aliquots (15 μL) were transferred in 10 µL of 250 mM NaOH, the precipitated protein was separated by centrifugation, D2O (600 μL) was added to supernatant and 31P NMR spectra were measured on a Bruker 500 UltraShieldTM. The experiment was performed in triplicate.
Molecular modelling
All molecular modelling studies were performed on a MacPro dual 2.66 GHz Xeon running Ubuntu 14LTS. The EcGadB structure was downloaded from the PDB data bank (http://www.rcsb.org/), PDB code 1PMM. We used a functional dimer of the hexameric enzyme, i.e., chains A and B. The aldimine PLP/Lys276 was bound at chain A, the PLP/L-Glu or L-Glu-γ-PH adducts were bound at chain B. The adducts structures were drawn by Maestro using the coordinates of the PLP and acetic acid (ACY) of the 1PMM structures. For the PLP-L-Glu external aldimine adduct the α-carboxy acid was deprotonated (carboxylate form) and the γ-carboxy acid was in a neutral from. For the PLP-L-Glu-γ-PH external aldimine adduct both the phosphinic and the α-carboxy acid groups were deprotonated. For both adducts, the pyridine and the aldimine nitrogen atoms were protonated. The MD was performed with the AMBER 12 suite61. For simulation, the protein was solvated in a periodic octahedron simulation box using TIP3P water molecules, providing a minimum of 10 Å of water between the protein surface and any periodic box edge. Ions were added to neutralise the charge of the total system. The solvent molecules and the ions were energy-minimised keeping the coordinates of the protein-ligand complex fixed (1000 cycle), and then the whole system was minimised keeping the adduct fixed (2000 cycle), then the all system was minimised without restraint (2000 cycle). Following minimisation, the entire system was heated to 298 K (2 ps). The production simulation was conducted at 298 K with constant pressure and periodic boundary condition (1 ns). Shake bond length condition was used (ntc = 2). Compounds were parametrised by Antechamber using BCC charges62,63. All the Asp, Glu and His residues of the binding site were titrated during the simulations keeping the protonation state fixed. The pH was kept at 4.5 by solvph = 4.5 and ntcnstph = 15,000 parameters in Amber production input file. Trajectories analysis was carried out by cpptraj programme64. The pictures reported in the manuscript were done with PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Fonda, M. Glutamate decarboxylase. Substrate specificity and inhibition by carboxylic acids. Biochemistry 11, 1304–1309 (1972).
Fonda, M. L. L-Glutamate decarboxylase from bacteria. Methods Enzymol. 113, 11–16 (1985).
Sukhareva, B. S. Amino acid decarboxylases. PyridoxaL Phosphate: Chemical, Biochemical and Medical Aspects Vol. 1B, 325–353 (Wiley, NY, 1986).
Gut, H. et al. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. EMBO J. 25, 2643–2651 (2006).
Lammens, T. M., De Biase, D., Franssen, M. C. R., Scott, E. L. & Sanders, J. P. M. The application of glutamic acid alpha-decarboxylase for the valorization of glutamic acid. Green Chem. 11, 1562–1567 (2009).
Kang, T. J., Ho, N. A. & Pack, S. P. Buffer-free production of gamma-aminobutyric acid using an engineered glutamate decarboxylase from Escherichia coli. Enzym. Microb. Technol. 53, 200–205 (2013).
Park, H. et al. Expression, immobilization and enzymatic properties of glutamate decarboxylase fused to a cellulose-binding domain. Int. J. Mol. Sci. 13, 358–368 (2012).
Ke, C. et al. Whole-cell conversion of l-glutamic acid into gamma-aminobutyric acid by metabolically engineered Escherichia coli. SpringerPlus 5, 591 (2016).
Xu, N., Wei, L. & Liu, J. Biotechnological advances and perspectives of gamma-aminobutyric acid production. World J. Microbiol. Biotechnol. 33, 64 (2017).
Tuck, C. O., Pérez, E., Horváth, I. T., Sheldon, R. A. & Poliakoff, M. Valorization of biomass: deriving more value from waste. Science 337, 695–699 (2012).
Zhang, J. et al. Metabolic engineering of Escherichia coli for the biosynthesis of 2-pyrrolidone. Metab. Eng. Commun. 3, 1–7 (2016).
Pennacchietti, E. et al. Mutation of His465 alters the pH-dependent spectroscopic properties of Escherichia coli glutamate decarboxylase and broadens the range of its activity toward more alkaline pH. J. Biol. Chem. 284, 31587–31596 (2009).
Grassini, G., Pennacchietti, E., Cappadocio, F., Occhialini, A. & De Biase, D. Biochemical and spectroscopic properties of Brucella microti glutamate decarboxylase, a key component of the glutamate-dependent acid resistance system. FEBS Open Bio 5, 209–218 (2015).
Vidal-Cros, A., Gaudry, M. & Marquet, A. Interaction of L-threo and L-erythro isomers of 3-fluoroglutamate with glutamate decarboxylase from Escherichia coli. Biochem. J. 229, 675–678 (1985).
Lupo, M. & Halpern, Y. S. Comparison of some physicochemical and catalytic properties of glutamate decarboxylase from various Escherichia coli K-12 sources. Biochim. Biophys. Acta 206, 295–304 (1970).
Homola, A. D. & Dekker, E. E. Decarboxylation of gamma-hydroxyglutamate by glutamate decarboxylase of Escherichia coli (ATCC 11246). Biochemistry 6, 2626–2634 (1967).
Jolles-Bergeret, B. & Charton, M. [Bacterial L-glutamate decarboxylases: their action on L homocysteinesulfinic acid and L-homocysteic acid]. Biochimie 53, 553–562 (1971).
Horsman, G. P. & Zechel, D. L. Phosphonate Biochemistry. Chem. Rev. 117, 5704–5783 (2017).
Metcalf, W. W. & van der Donk, W. A. Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78, 65–94 (2009).
Bunik, V. I. et al. Specific inhibition by synthetic analogs of pyruvate reveals that the pyruvate dehydrogenase reaction is essential for metabolism and viability of glioblastoma cells. Oncotarget 6, 40036–40052 (2015).
Peck, S. C. & van der Donk, W. A. Phosphonate biosynthesis and catabolism: a treasure trove of unusual enzymology. Curr. Opin. Chem. Biol. 17, 580–588 (2013).
Peck, S. C., Gao, J. & van der Donk, W. A. Discovery and biosynthesis of phosphonate and phosphinate natural products. Methods Enzymol. 516, 101–123 (2012).
Gill, H. S. & Eisenberg, D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 40, 1903–1912 (2001).
Abell, L. M. & Villafranca, J. J. Investigation of the mechanism of phosphinothricin inactivation of Escherichia coli glutamine synthetase using rapid quench kinetic technique. Biochemistry 30, 6135–6141 (1991).
Imai, S. et al. Studies on the biosynthesis of bialaphos (SF-1293). 6. Production of N-acetyl-demethylphosphinothricin and N-acetylbialaphos by blocked mutants of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. J. Antibiot. 38, 687–690 (1985).
Imai, S. et al. Studies on the biosynthesis of bialaphos (SF-1293). 4. Production of phosphonic acid derivatives, 2-hydroxyethylphosphonic acid, hydroxymethylphosphonic acid and phosphonoformic acid by blocked mutants of Streptomyces hygroscopicus SF-1293 and their roles in the biosynthesis of bialaphos. J. Antibiot. 37, 1505–1508 (1984).
Blodgett, J. A. et al. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat. Chem. Biol. 3, 480–485 (2007).
Seto, H. et al. Studies on the biosynthesis of bialaphos (SF-1293). 2. Isolation of the first natural products with a C-P-H bond and their involvement in the C-P-C bond formation. J. Antibiot. 36, 96–98 (1983).
Selvam, C., Goudet, C., Oueslati, N., Pin, J. P. & Acher, F. C. L-(+)−2-Amino-4-thiophosphonobutyric acid (L-thioAP4), a new potent agonist of group III metabotropic glutamate receptors: increased distal acidity affords enhanced potency. J. Med. Chem. 50, 4656–4664 (2007).
De Biase, D., Pennacchietti, E. & Giovannercole, F. Analoghi e derivati di amminoacidi dicarbossilici come antibatterici. ITA patent 102016000098005 (2016).
Froestl, W. et al. Phosphinic acid analogues of GABA. 1. New potent and selective GABAB agonists. J. Med. Chem. 38, 3297–3312 (1995).
Kuchenthal, C. H., Migenda, J., Polednia, M. & Maison, W. An improved protocol for the preparation of (S)-vinylglycine from (S)-methionine. Amino Acids 39, 443–448 (2010).
Xiao, Y., Lee, K. & Liu, P. Syntheses of the P-methylase substrates of the bialaphos biosynthetic pathway. Org. Lett. 10, 5521–5524 (2008).
Long, T. E. & Patel, S. K. Preparation of alkenes by mild thermolysis of sulfoxides. USA patent 20110230641 (2009).
Schwalbe, C. H. W., Freeman, S. & DasGupta, M. 2-Amino-2-carboxyethylphosphinic acid monohydrate. Acta Crystallogr. Sect. C 49, 1826–1828 (1993).
Baylis, E. K., Campbell, C. D. & Dingwall, J. G. 1-Aminoalkylphosphonous acids. Part 1. Isosteres of the protein amino acids. J. Chem. Soc. Perkin Trans. 1, 2845–2853 (1984).
Khomutov, M. A. et al. Convenient syntheses of phosphinic analogues of γ-aminobutyric- and glutamic acids. Rus. J. Bioorg. Chem. 42, 672–676 (2016).
Cassaigne, A., Lacoste, A. M. & Neuzil, E. [Non-enzymic transamination of aminophosphonic acids]. Biochim. Biophys. Acta 252, 506–515 (1971).
Lacoste, A. M., Mansour, S., Cassaigne, A. & Neuzil, E. Effect of phosphonic analogues of glutamic acid on glutamate decarboxylase. Experientia 41, 643–644 (1985).
Mongan, J., Case, D. A. & McCammon, J. A. Constant pH molecular dynamics in generalized Born implicit solvent. J. Comput. Chem. 25, 2038–2048 (2004).
Capitani, G. et al. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 22, 4027–4037 (2003).
Tramonti, A., John, R. A., Bossa, F. & De Biase, D. Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur. J. Biochem. 269, 4913–4920 (2002).
Scott, E. M. & Jakoby, W. B. Soluble gamma-aminobutyric-glutamic transaminase from Pseudomonas fluorescens. J. Biol. Chem. 234, 932–936 (1959).
Jakoby, W. B. & Scott, E. M. Aldehyde oxidation. III. Succinic semialdehyde dehydrogenase. J. Biol. Chem. 234, 937–940 (1959).
Damiano, M. A. et al. Glutamate decarboxylase-dependent acid resistance in Brucella spp.: distribution and contribution to fitness under extremely acidic conditions. Appl. Environ. Microbiol. 81, 578–586 (2015).
De Biase, D., Tramonti, A., John, R. A. & Bossa, F. Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr. Purif. 8, 430–438 (1996).
Laber, B. & Amrhein, N. Metabolism of 1-aminoethylphosphinate generates acetylphosphinate, a potent inhibitor of pyruvate dehydrogenase. Biochem. J. 248, 351–358 (1987).
Hyvönen, M. T. et al. Enantiomers of 3-methylspermidine selectively modulate deoxyhypusine synthesis and reveal important determinants for spermidine transport. ACS Chem. Biol. 10, 1417–1424 (2015).
Faleev, N. G. et al. Methionine gamma-lyase: mechanistic deductions from the kinetic pH-effects. The role of the ionic state of a substrate in the enzymatic activity. Biochim. Biophys. Acta 1794, 1414–1420 (2009).
Faleev, N. G. et al. Interaction of tyrosine phenol-lyase with phosphoroorganic analogues of substrate amino acids. Eur. J. Biochem. 267, 6897–6902 (2000).
Schulz, A., Taggeselle, P., Tripier, D. & Bartsch, K. Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: isolation and characterization of a phosphinothricin-specific transaminase from Escherichia coli. Appl. Environ. Microbiol. 56, 1–6 (1990).
Vrtis, J. M., White, A. K., Metcalf, W. W. & van der Donk, W. A. Phosphite dehydrogenase: a versatile cofactor-regeneration enzyme. Angew. Chem. 41, 3257–3259 (2002).
Nemeria, N. et al. The 1′,4′-iminopyrimidine tautomer of thiamin diphosphate is poised for catalysis in asymmetric active centers on enzymes. Proc. Natl Acad. Sci. USA 104, 78–82 (2007).
Matziari, M. & Yiotakis, A. Shortcut to fmoc-protected phosphinic pseudodipeptidic blocks. Org. Lett. 7, 4049–4052 (2005).
Khomutov, A. R. et al. Synthesis of hydroxylamine analogues of polyamines. Tetrahedron 52, 13751–13766 (1996).
Vogel, H. J. & Bonner, D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106 (1956).
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997).
Mathlouthi, A., Pennacchietti, E. & De Biase, D. Effect of temperature, pH and plasmids on in vitro biofilm formation in Escherichia coli. Acta Nat. 10, 129–132 (2018).
Wiegand, I., Hilpert, K. & Hancock, R. E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
CLSI. Methods for dilution antimicrobial susceptibility tests for Bacteria that grow aerobically; Approved standards-tenth edition. M07-A10 Vol. 35 (Clinical and Laboratory Standards Institute, Wayne, Pennsylvania, 2015).
Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006).
Roe, D. R. & Cheatham, T. E. 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Acknowledgements
This work was supported by Sapienza University of Rome (Project RM11916B861B9985) to D.D.B., by Academy of Finland (Grant 315487) to J.V. and by the Russian Foundation of Basic Research (Grant No. 17–00–00395) to A.K. The authors thank Dr. A.O. Chizhov (N.D. Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Moscow) for measurement of mass spectra.
Author information
Authors and Affiliations
Contributions
D.D.B. and A.K. conceived the study and interpreted the results. D.D.B., F.C., E.P., F.G. and A.K. performed experiments and analysed the data from the relevant experiments. J.V. and A.K. interpreted NMR spectra. A.C. performed the molecular modelling. D.D.B. and A.K. wrote the paper. E.P., F.G., A.C. and J.V. read the paper and provided feedback.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Biase, D., Cappadocio, F., Pennacchietti, E. et al. Enzymatic kinetic resolution of desmethylphosphinothricin indicates that phosphinic group is a bioisostere of carboxyl group. Commun Chem 3, 121 (2020). https://doi.org/10.1038/s42004-020-00368-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-020-00368-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.