Abstract
Long non-coding RNAs (lncRNAs) are emerging important epigenetic regulators in metabolic processes. Whether they contribute to the metabolic effects of vertical sleeve gastrectomy (VSG), one of the most effective treatments for sustainable weight loss and metabolic improvement, is unknown. Herein, we identify a hepatic lncRNA Gm19619, which is strongly repressed by VSG but highly up-regulated by diet-induced obesity and overnight-fasting in mice. Forced transcription of Gm19619 in the mouse liver significantly promotes hepatic gluconeogenesis with the elevated expression of G6pc and Pck1. In contrast, AAV-CasRx mediated knockdown of Gm19619 in high-fat diet-fed mice significantly improves hepatic glucose and lipid metabolism. Mechanistically, Gm19619 is enriched along genomic regions encoding leptin receptor (Lepr) and transcription factor Foxo1, as revealed in chromatin isolation by RNA purification (ChIRP) assay and is confirmed to modulate their transcription in the mouse liver. In conclusion, Gm19619 may enhance gluconeogenesis and lipid accumulation in the liver.
Similar content being viewed by others
Introduction
Vertical sleeve gastrectomy (VSG) is currently one of the most popular and efficacious bariatric surgeries for sustainable weight loss and the subsequent improvement of diabetes and other obesity-related complications1,2. Previously, we and other groups have reported several molecular mechanisms that contribute to the effects of VSG3,4,5,6,7. Briefly, we reported that the membrane-bound G protein-coupled bile acid receptor TGR5 was required to maintain the metabolic effects of VSG in mice5,8. The expression of TGR5 was increased significantly by VSG to improve glucose control and increase energy expenditure. Recently, we revealed that the intestinal levels of bile acids were significantly reduced by VSG, thereby restricting fat absorption in the intestine6,9. However, the molecular mechanism underlying the metabolic improvement of VSG is still largely unknown. A better understanding of the molecular mechanism of VSG may lead to the development of novel therapeutic treatments for obesity and avoid invasive surgery10,11.
Besides their protein-coding counterparts, long non-coding RNAs (lncRNAs) have been reported to play important roles in metabolic regulation. For example, activation of the liver X receptors (LXR) can robustly induce the expression of a hepatic lncRNA LeXis, which then affects the expression of genes involved in cholesterol biosynthesis and alters the cholesterol levels12. Cholesterol is also reported to induce other liver lncRNAs, such as CHROME, to promote cholesterol efflux and high-density lipoprotein (HDL) biogenesis by binding to several functional microRNAs13. Moreover, the lncRNA lncOb, which is found to be important for leptin expression, is repressed in diet-induced obese mice and can lead to a leptin-responsive form of obesity14. However, little is known whether lncRNAs mediate the metabolic effects of bariatric surgery.
In this study, we took advantage of our well-established VSG mouse models to screen for candidate lncRNAs whose hepatic expression was responsive to VSG operation in mice. Among them, we identified and characterized the functions of a hepatic nuclear lncRNA Gm19619, which is repressed by VSG in mice. Interestingly, Gm19619 was strongly induced by both diet-induced obesity and overnight-fasting. Forced transcription of Gm19619 by AAV in the mouse liver significantly enhanced hepatic gluconeogenesis. In contrast, AAV-CasRx mediated knockdown of Gm19619 in HFD-fed mice profoundly improved both hepatic glucose and lipid metabolism. We further demonstrated that Gm19619 may promote gluconeogenesis by activating the Foxo1-G6pc/Pck1 pathway, and facilitate lipid accumulation by inhibiting the leptin receptor (Lepr) signaling pathway.
Results
VSG strongly suppressed the transcription of Gm19619
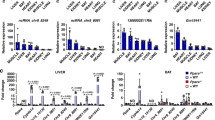
In our previous studies, we had succeeded in performing the VSG operation on HFD-induced obese mice5,6. The results showed that VSG could induce a significant weight loss in mice when compared with sham-operated mice. Blood glucose regulation was also strikingly improved in VSG-operated mice. The RNAseq result confirmed the repression of several key genes involved in lipogenesis, such as Fasn, Scd1, Cd36, and Srebf1, and two gluconeogenesis marker genes G6pc and Pck1 (Fig. 1a). These data suggested that the VSG on mice faithfully recapitulates its metabolic effects in human patients. Due to the emerging important roles of lncRNA in metabolism, we profiled the differentially transcribed liver lncRNAs between VSG- and sham-operated mice as well. The results showed around 1900 lncRNAs were mapped to the mouse genome and 40 lncRNAs were found to be significantly changed (fold change >1.67 and p < 0.05) after VSG operation in mice (Fig. 1a). Among them, Gm19619 was verified by qPCR to be a top candidate that was strongly downregulated in the livers of VSG-operated mice. Moreover, we further confirmed that the hepatic expression level of Gm19619 in lean mice was much lower compared to that in diet-induced obese mice (Fig. 1b). Additionally, in the overnight-fasted mice, fasting strongly elevated the transcriptional level of Gm19619 (Fig. 1c). In mouse immortal hepatocytes AML12 cell line, mimicking the activation of glucagon pathway by forskolin treatment strongly elevated the transcription of Gm19619 (Fig. 1d). All these results consistently indicate that Gm19619 is a highly responsive lncRNA to VSG and metabolic conditions, which highlights its potential function in regulating hepatic metabolism.
a Heatmap showing the several protein-coding metabolic genes involved in lipogenesis (Fasn, Scd1, Cd36, and Srebf1) and gluconeogenesis (G6pc and Pck1), and the top 40 differentially transcribed lncRNAs with a fold change over 1.67 (p < 0.05) of the counts in FPKM in the RNAseq data of sham or vertical sleeve gastrectomy (VSG) operated mice livers (n = 3). b The relative level of different lncRNAs in lean, sham, or VSG-operated mice liver by real-time PCR (n = 3). c, d The relative RNA level of Gm19619, G6pc, and Pck1 in overnight-fasted (16 h) (n = 5) and re-fed (4 h) (n = 7) mice liver (c), and in murine immortal hepatocytes AML12 cells after treatment of 10 μM forskolin for 7, 12, and 18 h (n = 3) (d). e The full length of Gm19619 RNA was determined by RT-PCR using mouse liver cDNA after its 5′ and 3′ sequence was confirmed by RACE-PCR. f Real-time PCR determined the distribution of Gm19619 RNA in nucleus and cytosol (n = 3). Rnu2 and Gapdh RNA served as the marker of the nucleus and cytosol, respectively. Two-tailed unpaired t-test. Error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
For further study, the comparative genomics method PhyloCSF was first applied to check that Gm19619 only possessed a barely translational potential15. The data from the FANTOM5 database indicated that Gm19619 was specifically transcribed in mouse liver but no other tissues (Supplementary Fig. 1)16. Both the 5′ and 3′ ends of the Gm19619 transcript were determined by the rapid amplification of cDNA ends PCR (RACE-PCR)17,18. By using the primers targeting both ends of Gm19619 full-length RNA and mouse hepatic cDNA as templates for PCR, we confirmed that Gm19619 has only one isoform around 1600 nt in mouse liver with a 3′ poly(A) tail (Fig. 1e and Table 1). Because the cellular location is highly correlated with the function of lncRNA, the cytosolic and nuclear RNA in mouse primary hepatocytes were fractionated respectively for quantification. Most of the Gm19619 RNA was confirmed to be in the nucleus, indicating its potential regulatory function in chromatin gene transcription (Fig. 1f).
Forced expression of Gm19619 enhanced hepatic gluconeogenesis
Because fasting significantly upregulated the transcription of Gm19619, we asked whether the forced expression of Gm19619 RNA would affect the hepatic glucose metabolism in chow-fed mice. To this end, the adeno-associated viruses (AAV) for liver-specific TBG promoter-driven Gm19619 transcription was constructed, purified, and intravenously injected into mice. GFP was used as a negative control (Fig. 2a). As expected, the AAV strongly increased the hepatic Gm19619 RNA level (Fig. 2b). In addition, fasting can further elevate the RNA level of Gm19619 when compared with the re-fed group in both AAV-Gm19619 and AAV-GFP mice (Fig. 2b). The mRNA levels of both G6pc and Pck1 were significantly upregulated after overnight fasting. Interestingly, the forced transcription of Gm19619 could further promote the increase of both G6pc and Pck1 mRNAs (Fig. 2b). The western blot assays indicated that while there was no significant difference between the expression levels of these two genes in fasted and re-fed group mice, Gm19619 significantly increased the expression levels of both G6PC and PCK1 in both groups of mice (Fig. 2c). Consistent with the expression of these genes, the fasting blood glucose concentration was significantly upregulated after Gm19619 forced transcription (Fig. 2d). Glucose tolerance test (GTT) and insulin tolerance test (ITT) results further confirmed that elevation of Gm19619 transcription impaired the glucose metabolism and insulin sensitivity in mice (Fig. 2e, f), while the pyruvate tolerance test (PTT) result verified that Gm19619 could remarkably enhance gluconeogesis in the liver (Fig. 2g), although both the body weight and liver/body weight ratio were comparable between these mice (Supplementary Fig. 2a, b). H&E staining showed no detectable morphology changes in either the liver, inguinal white adipose tissue (iWAT), or epididymal white adipose tissue (eWAT) after Gm19619 forced transcription (Supplementary Fig. 2c). The mRNA level of lipogenic genes, including Fasn and Scd1, remained unchanged (Supplementary Fig. 2d). These results suggest that Gm19619 may promote hepatic gluconeogenesis but not affect lipid metabolism under normal conditions.
a Schema of the AAV vector for hepatic Gm19619 transcription under the liver-specific TBG promoter and the experimental timeline. GFP was used as the control. b The relative RNA level of Gm19619, G6pc, and Pck1 in AAV-Gm19619 (n = 9) or AAV-GFP (n = 7) infected mice liver after 16 h fasting and 4 h refeeding by real-time PCR. c Western blot analysis of protein levels of G6PC and PCK1 in AAV-Gm19619 and AAV-GFP mice liver (n = 3). d The blood glucose level of AAV-Gm19619 (n = 18) and AAV-GFP (n = 11) mice after overnight fasting. e–g GTT (e) (n = 11), ITT (f) (n = 11), and PTT (g) (n = 8) assay of the AAV-Gm19619 and AAV-GFP mice. Two-tailed unpaired t-test. Error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Knockdown of Gm19619 in the HFD-fed mice inhibited the gluconeogenesis
The highly elevated levels of Gm19619 in both obese mice and overnight-fasted mice and its potential role in promoting hepatic gluconeogenesis suggest that it may also be involved in the pathogenesis of diet-induced obesity and diabetes. To determine its potential function in obesity, we constructed the AAV-based vector containing the CasRx gene with concatenated nuclear localization signal peptides to specifically knockdown Gm19619 lncRNA in the nucleus. Three concatenated sgRNAs targeting Gm19619 spaced by DR36 sequence were inserted and transcribed under the human U6 promoter (Fig. 3a). The quantitative PCR results indicated the intravenously injected AAV efficiently silenced the hepatic Gm19619 transcription in the HFD-fed mice (Fig. 3b). As expected, knockdown of Gm19619 RNA significantly lowered the mouse fasting blood glucose level (Fig. 3c), improved the glucose tolerance (GTT assay) and the insulin resistance (ITT assay) of HFD-fed mice (Fig. 3d, e). Moreover, the PTT assay further confirmed that Gm19619 knockdown repressed gluconeogenesis in mice (Fig. 3f). Accordingly, the mRNA level of the Pck1 gene was significantly decreased, although G6pc remained unchanged (Fig. 3b). The protein levels of G6PC and PCK1 were found to be consistent with their transcriptional changes (Fig. 3g). These results indicated that knockdown of Gm19619 could significantly ameliorate glucose intolerance and insulin resistance in HFD-fed mice.
a Schema of the AAV-EFS-CasRx vector for Gm19619 knockdown and the experimental timeline. b The relative RNA levels of hepatic Gm19619, G6pc, and Pck1 in Gm19619 knockdown (CasRx-19×3) (n = 10) and control (CasRx-ctrl) (n = 7) mice. c The fasting blood glucose levels in CasRx-19×3 mice (n = 10) and CasRx-ctrl (n = 7) mice. d–f The GTT (d), ITT (e), and PTT (f) assay of CasRx-19×3 (n = 10, PTT n = 8) and CasRx-ctrl (n = 7) mice. g The western blot results of G6PC and PCK1 in CasRx-19×3 and control group mice livers (n = 3). Two-tailed unpaired t-test. Error bars represent the SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
Knockdown of Gm19619 in the HFD-induced obese mice improved lipid metabolism
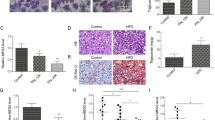
Although knockdown of Gm19619 showed no significant effect on the body weight when compared with the control group (Fig. 4a), H&E staining and oil red o staining showed smaller droplets and less lipid accumulation in Gm19619 knockdown group (Fig. 4b), suggesting its role in hepatic lipid metabolism under obesity condition. The hepatic triglycerides (TG) levels were confirmed to be strongly decreased after Gm19619 knockdown (Fig. 4c), while the amount of serum non-esterified fatty acid (NEFA) was significantly upregulated (Fig. 4d). These changes were accompanied by the decreased mRNA levels of several lipogenesis genes such as Srebf1, Fasn, Scd1, and the fatty acid absorption receptor Cd36 (Fig. 4e). Moreover, the transcription of several genes mediating lipolysis and fatty acid beta-oxidation, including Acsl1, Pnpla2, Lipe, Cpt1a, and Cpt1b, were elevated by Gm19619 knockdown (Fig. 4f), indicating Gm19619 can promote lipogenesis and inhibit lipolysis pathways in the HFD-fed mice liver.
a The body weight of Gm19619 knockdown (CasRx-19×3) (n = 11) and control (n = 8) mice. b Representative images of liver sections by H&E and oil red o staining, respectively. Scale bar, 100 μm. c The serum level of triacylglycerol (TG) in CasRx-19×3 (n = 11) and CasRx-ctrl (n = 6) mice. d The serum level of NEFA in CasRx-19×3 (n = 11) and CasRx-ctrl (n = 8) mice. e The relative mRNA levels of some lipogenesis genes and Cd36 in CasRx-19×3 (n = 11) and CasRx-ctrl (n = 8) mice. f The relative mRNA levels of several lipolysis and beta-oxidation genes in CasRx-19×3 and CasRx-ctrl mice (n = 8). Two-tailed unpaired t-test. Error bars represent the SEM. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
Gm19619 downregulated hepatic metabolic signaling pathways
To investigate the potential mechanism by which Gm19619 regulates hepatic metabolism, we first tested if the transcriptional level of its two nearby genes Areg (Amphiregulin) and Btc (Betacellulin), was correlated with Gm19619 RNA, as many lncRNAs function by modulating their nearby gene level19. The real-time PCR data indicated that Areg was barely transcribed in the liver and not correlated with Gm19619 (Supplementary Fig 3a), while the Btc RNA level was much higher in sham mice liver than VSG mice liver, which was like Gm19619. However, the forced transcription of Gm19619 did not affect the transcription of Btc in the liver (Supplementary Fig. 3b), indicating that Gm19619 may not function by regulating its nearby gene expression.
For the further mechanistic study, we performed chromatin isolation by RNA purification (ChIRP) assay by using freshly fixed primary murine hepatocytes from wild-type mice. Fourteen probes with antisense to different regions of Gm19619 RNA were divided into two sets (odd- and even-numbered) for the assay (Fig. 5a and Supplementary Table 1). The pull downed chromatin DNA was sequenced and mapped to the mouse genome20. The analysis of the mapped DNA sequences indicates that most of Gm19619 RNA bind to the distal intergenic regions and introns, while 9% of the binding region was within the promoter region (Fig. 5b). The pathway enrichment analysis of genes showing interactions with Gm19619 revealed that leptin receptor signaling was among the top potential pathways affected by Gm19619 (Fig. 5c, d). While Gm19619 showed binding to its own genomic locus (Fig. 5e), ChIRP-seq also revealed the Gm19619 binding to the upstream region and genomic locus of Lepr (Fig. 5f), as well as Plcg1, Adcy8, Prkar2a, and Npy (Supplementary Fig. 4). The function of leptin receptor signaling in obesity pathogenesis has been well reported, in part supported by the striking metabolic phenotype exhibited by the db/db mouse which carries Lepr mutation21.
a The second structure of Gm19619 RNA with minimal free energy as predicted on the RNAfold website. The locations of the 14 ChIRP probes were shown. b The genomic distribution of ChIRP pulldown DNA fragments with a pileup over 5 and longer than 200 bp. c Pathway enrichment analysis of genes with surrounding regions bound by Gm19619 identified from ChIRP-seq. The gene list from overlapping peaks was used to run KEGG pathway analysis with IPA (Ingenuity Pathway Analysis). d The Circos plot showing the ChIRP enriched genes within the leptin receptor pathway. e, f, k ChIRP-seq signals indicate Gm19619 binding along its own exon locus (e), the leptin receptor Lepr (f), and the Foxo1 gene locus (k). H3K27ac, H3K4me1, and H3K4me3 signals from ENCODE mouse liver ChIP-seq data were shown as control, mm10 as the reference genome. g The relative mRNA levels of Lepr, Plcg1, and Foxo1 in Gm19619 forced expressed mice (n = 5–10). h Western blot analysis of protein levels of LEPR, FOXO1, and CREB1 in AAV-Gm19619 and AAV-GFP mice liver (n = 3). i The relative mRNA levels of Lepr, Plcg1, and Foxo1 in Gm19619 knockdown mice (n = 6–7). j The western blotting of LEPR, FOXO1, and CREB1 in the livers of CasRx-19×3 and control mice (n = 3). Two-tailed unpaired t-test. Error bars represent the SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
QPCR data showed the transcriptional level of Lepr was strikingly induced in overnight-fasted mice when compared with the re-fed mice (Fig. 5g), and AAV-Gm19619 significantly reduced the fasting-induced Lepr mRNA level. However, western blotting showed that fasting suppressed the long active isoform of LEPR (~125 kDa) expression, and AAV-Gm19619 did not affect the LEPR expression in both fasting and feeding status (Fig. 5h), consistent with the negligible effect of Gm19619 on hepatic lipid metabolism in chow-fed mice (Supplementary Fig. 2). In contrast, the Gm19619 knockdown highly upregulated both the mRNA and protein levels of LEPR in HFD-fed mice (Fig. 5i, j). Similarly, the transcriptional repression of Plcg1 by fasting was interrupted in AAV-Gm19619 mice (Fig. 5g); although the knockdown of Gm19619 did not affect the mRNA level of Plcg1 (Fig. 5i). These results suggest that Gm19619 may repress the leptin receptor pathway in the liver, which contributed to its regulation of hepatic lipid metabolism mainly under the obese condition.
ChIRP data also identified that Gm19619 could bind to the promoter region and genomic locus of Foxo1, a key transcription factor (TF) in gluconeogenesis (Fig. 5k), suggesting that Foxo1 may also be a potential downstream effector of Gm19619 to promote the gluconeogenesis in the liver. Consistent with a recent report22, the protein expression of FOXO1 was found to be repressed in fasted mouse liver when compared with that in the re-fed mouse liver (Fig. 5h). In the fasted mice, AAV-Gm19619 significantly increased both the mRNA and protein levels of hepatic Foxo1, but not Creb1 (Fig. 5g, h). On the other hand, Gm19619 knockdown in HFD-fed mice repressed the expression of FOXO1 (Fig. 5i, j), which might contribute to the improved glucose metabolism in HFD-fed mice.
In summary, our data show that VSG in mice significantly represses the transcription of hepatic Gm19619, while both obesity and fasting induce its upregulation. Gm19619 lncRNA may promote gluconeogenesis by activating the Foxo1 pathway, and facilitate lipogenesis and repress lipolysis by inhibiting the hepatic leptin receptor signaling (Fig. 6).
Discussion
VSG is currently one of the most common and effective treatments for the sustained weight loss and remission of diabetes, as well as other comorbidities such as nonalcoholic fatty liver disease (NAFLD). However, it is still an invasive surgery with low mortality probability and may not be suitable for some specific groups of people. A better understanding of the mechanism underneath the beneficial effect of VSG or other bariatric surgeries may lead to the development of less invasive and cost-effective therapeutic approaches. In this study, we used VSG-operated mice and fasting/refeeding mice as models and found that the obesity and fasting-induced hepatic lncRNA Gm19619 was highly suppressed after VSG operation. Forced transcription of Gm19619 activated hepatic gluconeogenesis, while AAV-CasRx mediated in vivo hepatic knockdown significantly improved the glucose and lipid metabolism in HFD-fed mice.
Mechanically, Gm19619 may affect several metabolic signaling pathways that regulate hepatic glucose and lipid metabolism. Among them, the hepatic leptin receptor signaling pathway is a major downstream mediator of Gm19619. Leptin receptor signaling is particularly important in obesity pathogenesis. Some recent studies showed that mere ablation of hepatic leptin receptors could promote lipid accumulation in the liver and produce more enlarged triglyceride-rich VLDL particles23,24. Our results confirmed that the leptin receptor pathway was regulated by Gm19619, particularly in the HFD-fed mice, indicating that Gm19619 might promote lipid accumulation by repressing the leptin receptor pathway in the liver under the obese condition. Surprisingly, the circulating leptin level was found to be reduced after VSG25, and yet VSG can still reduce fat mass and improve glucose tolerance in ob/ob mice in the absence of the leptin signaling26. In this regard, in addition to the repression of Gm19619 in the liver, other signaling pathways may also have a significant impact on lipid metabolism after the VSG surgery.
Moreover, a recent study reported that activation of hepatic leptin signaling improved hyperglycemia by degrading MARK phosphatase-3 (also known as dual specificity phosphatase 6, DUSP6) and subsequent repressing Foxo1-mediated gluconeogenesis pathway27, indicating Gm19619 may promote the hepatic gluconeogenesis by repressing the hepatic leptin signaling. Indeed, we found that Gm19619 also bound to the promoter of the Foxo1 and activated its expression to modulate the hepatic glucose metabolism. Thus, the downregulation of Gm19619 by VSG may contribute to the improved hepatic glucose metabolism in part through the Foxo1 pathway. The distinct consequence of Gm19619 binding in different genes (e.g., Lepr vs Foxo1) will warrant future investigation.
Besides Gm19619, we identified other hepatic lncRNAs differentially expressed after VSG surgery. For example, a lncRNA Gm10680, which was also significantly repressed by VSG, has been reported to be involved in hepatic lipid metabolism. Knockdown of this lncRNA reduced the nearby APOA4 gene expression and led to reduced levels of plasma triglyceride and total cholesterol in ob/ob mice28. It is likely that Gm10680 and other lncRNAs involved in the various metabolic pathways can collectively lead to metabolic improvement after VSG surgery. In addition to the liver lncRNAs, lncRNAs such as GAS5 in adipose tissues, small intestine, and other tissues may also contribute to the overall metabolic improvement by VSG29,30.
LncRNAs can recruit RNA-binding proteins and transcription factors to regulate downstream gene function. To identify potential binding proteins of Gm19619, we analyzed the binding motifs and TF binding sites enriched in the binding sequences of Gm19619. This analysis revealed that Gm19619 could likely interplay with several TFs, including SREBF1, SREBF2, FXR, LXR, and HNF4α, to regulate hepatic glucose and lipid metabolism (Supplementary Fig. 5). Moreover, the prediction of potential TFs that may bind to and regulate Gm19619 promoter activity (by the JASPAR31) showed that Gm19619 may be regulated by CREB1, PPARα, FXR, and additional TFs known to regulate glucose metabolism in fasted and fed states (Supplementary Fig. 6)32. A more comprehensive investigation of the metabolic modulation of Gm19619 will be our future endeavor.
We used AAV-CasRx to knock down the hepatic RNA of Gm19619. Compared with CRISPR-Cas9 mediated chromatin DNA disruption or CRISPRi mediated transcriptional repression33, CasRx will only bind and cut RNA34, without any interaction with chromatin DNA. CRISPR-Cas9 with a single guide RNA for gene knockout does not work for lncRNA, as a small insertion or deletion may not affect the function of most lncRNAs. Therefore, pair of sgRNAs is always applied to knock out a large DNA fragment in the lncRNA locus to totally abolish its transcription35. However, this technique may fail to dissect if the disrupted function of lncRNA is due to the transcript itself or the DNA elements, such as enhancers on the chromosome. Currently, there are no CasRx transgenic mice available. It can be anticipated that the development of this kind of mice will facilitate the in vivo studies of lncRNAs in the future.
The discovery of lncRNA expands our knowledge of the complexity of vertebrate genomes, but the low sequence conservation among varied species may be a barrier to the understanding of relevant human physiology and disease research. One approach to address this problem may be the use of humanized mice. For example, Ruan et al. used a humanized TK-NOG mouse model where the liver was repopulated with human hepatocytes to investigate the function of non-conserved human hepatic lncRNAs associated with NAFLD and metabolic homeostasis. They managed to identify LINC01018 as an obesity-associated lncRNA, and hLMR1 promotes the transcription of cholesterol biosynthetic genes36,37. Further effort should be made to test if such humanized mouse models can be used for bariatric surgery studies, which may better recapitulate the situation in human patients. Another alternative approach will be using the human lncRNA knock-in mouse models, although many challenges remain to be solved38.
Methods
Animals
Wild-type C57BL/6 male mice (4–8 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME). All animal procedures were approved by the City of Hope Institutional Animal Care and Use Committee (IACUC protocol: #14031) and conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Mice were housed in a 12-h light/dark cycle in a temperature-controlled room (25 °C) with a relative humidity of 50%. Animals were given ad libitum access to water and a regular chow diet in a temperature-controlled environment.
Primary hepatocytes isolation
Primary hepatocytes were isolated by two-step methods. The anesthetic mouse was perfused with 50 mL 1× DPBS without Ca2+ and with 5 mM EDTA through a portal vein at a speed of 5 mL/min, then 25 mg of collagenase IV (Worthington) dissolved in 50 mL of HBSS buffer was perfused. The liver was harvested and rid of the gallbladder. The hepatocytes were shaken off by tweezers into DMEM with 10% FBS and antibiotics and collected by centrifuge at 50 × g for 3 min. The cells were then washed twice with DMEM with 10% FBS. The purified primary hepatocytes were immediately seeded into a collagen-pretreated six-well plate or used for other experiments.
Primary hepatocytes cytosolic and nuclear RNA isolation
The nucleus and cytosol of the hepatocytes were isolated by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher) according to the vendor’s instruction. SUPERase•In RNase inhibitor was added into the separation buffers to prevent RNA degradation. The RNA was directly isolated by the Direct-zol RNA kit (Zymo Research) and eluted using the same volume of RNase-free water. About 1 ug cytoplasmic RNA and the same volume of nuclear RNA was reversely transcribed respectively by using the high-capacity cDNA reverse transcription kit from Thermo Fisher.
RACE-PCR
The nucleus RNA isolated from murine hepatocytes was used for the 5′ and 3′ ends of Gm19619 determination by the RACE-PCR according to the published protocols17,18. Briefly, for the 5′ RACE, the primer RA5-1 (5′-CTCAGCCAATGATCTCCAAC-3′) was used for reverse transcription, then the RNA template was degraded by RNase H and poly(A) tail was added to first-strand cDNA products by the terminal deoxynucleotidyl transferase. Three primers RA5-2 (5′-GTGACATAGGTCCTTTGGC-3′), Qdt (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGCTTTTTTTTTTTTTTTTT-3′), and Qouter (5′-CCAGTGAGCAGAGTGACG-3′) was used for the first round of the PCR. The primers RA5-3 (5′-GCTTGAAGACTCCTTGCTG-3′) and Qinner (5′-GAGGACTCGAGCTCAAGC-3′) were used for the second round of PCR.
For the 3′ RACE, the primer Qdt was used for the RNA reverse transcription, then primer Gm19619_QF (AAGCATCAGAGTCCAGGGTC) and Qouter were used for the first round of PCR. The primer 3RA22-F(5′-ACAGCACTTGGCCTGCAAAC-3′) and Qinner were used for the second round of PCR. The amplicons of these PCR were gel purified and sent for sanger sequencing.
Based on the sequencing results, the primers Gm19619_nFo (5′-CCCACCCAGGAACAGAGACTTTA-3′) and Gm19619_nRo (5′-AGTGGCATACTGTCATATATATTAGAACAAA-3′) was used for the full-length PCR amplification.
AAV-TBG-Gm19619 for lncRNA Gm19619 transcription in chow-fed mice
The sequence of Gm19619 was amplified by the primers Gm19619_NotI_F (5′-aGCGGCCGCccacccaggaacagagact-3′) and Gm19619_Esp3I_R (5′-tCGTCTCaGATCagtggcatactgtcatatatattag-3′), digested and replaced the eGFP sequence in pAAV-TBG-eGFP (Addgene #105535). The plasmid was sequenced and then used for AAV packing in 293 T cells according to the AAV protocols at the Addgene website and our previous report39. Eight-week-old C57BL/6 mice were received once intravenous injections of 5 × 1011 genome copies of AAV-TBG-Gm19619, or AAV-TBG-eGFP for control. Animals were fed with the standard chow ad libitum. The mice were euthanized after being fasted for 16 h or a 16 h fasting followed by 4 h refeeding.
AAV-CasRx mediated knockdown of lncRNA Gm19619 in HFD mice
The plasmid of pAAV-EFS-CasRx was constructed by cloning the EFS promoter and RfxCas13d from the pLentiRNACRISPR_005 plasmid (Addgene #138147) and replaced the TBG promoter and SaCas9 gene in px602 plasmid (Addgene #61593). The DR30 sgRNA cassette for CasRx sgRNA cloning was used to further replace the Sa gRNA scaffold. Five individual sgRNA candidates targeting Gm19619 was inserted after the DR30 respectively and verified their knockdown efficacy in 293 T cells against the co-transfected pCMV-Gm19619 plasmid by QPCR. Then three sgRNAs were chosen to be concatenated and spaced by DR36 sequences to construct the pAAV-EFS-CasRx-19×3 plasmid for AAV packaging. Four-week-old C57BL/6 mice were fed with HFD for 3 weeks, then received once intravenous injections of 5 × 1011 genome copies of AAV-CasRx-19×3, as well as AAV-CasRx-GFP for control and continued on HFD until euthanization.
Glucose tolerance test (GTT)/pyruvate tolerance test (PTT)
Mice were fasted for 14 h prior to GTT or PTT. An AlphaTRAK 2 Blood Glucose Monitoring System was used to measure blood glucose from blood collected by tail nick. After the initial blood glucose reading, mice were intraperitoneally injected with glucose solution at 2 g/kg of body weight, or pyruvate solution at 2.5 g/kg of body weight40. Then, blood glucose was measured at 15, 30, 60, 90, and 120 min after glucose or pyruvate injection.
Insulin tolerance test (ITT)
Mice were fasted for 6 h prior to ITT. Blood glucose level was measured from blood collected by tail prick with an AlphaTRAK 2 Blood Glucose Monitoring System. After the initial blood glucose reading (Time 0 min), mice were injected with insulin intraperitoneally at 0.75 U/kg of body weight. Blood glucose was measured at 15, 30, 60, and 120 min after insulin injection.
Serum analysis
Serum ALT (alanine transaminase) and AST (aspartate transaminase) levels were measured using the EnzyChrom™ Alanine Transaminase Assay Kit (Bioassay, EALT100, Hayward, CA) and the EnzyChrom™ Aspartate Transaminase Assay Kit (Bioassay, EASTR-100, Hayward, CA). Serum levels of non-esterified FFA (NEFA) were measured using a NEFA Assay kit (Wako, TK036, Neuss, Deutschland). Hepatic triglyceride levels were measured using the L-Type Triglycerides M Kit (Wako, TK006, Neuss, Deutschland).
Hematoxylin and eosin (H&E) and Oil Red O staining
After mice were euthanized, their tissues were immediately removed, immersed in cold PBS, and fixed in 10% formalin solution for 24–48 h at 4 °C. For H&E staining, the samples were then incubated in 70% ethanol for 12 h, embedded in paraffin, and sectioned into 5 μm sections. For Oil Red O staining of the liver sections, the tissues were fixed in 4% paraformaldehyde, then incubated in 30% sucrose for 12 h and embedded in Tissue-Tek® O.C.T. Compound (Sakura Finetek, Torrance, CA, USA). Serial sections (10 μM) were made and stained with 0.5% Oil Red O for 10 min and counter-stained with hematoxylin. The red lipid droplets were visualized by microscopy.
Western blot
The liver lysate samples of 30 μg per lane of gel were used for the SDS-PAGE and western blot. The used antibodies are listed below: G6PC (1:500), Sigma Aldrich, HPA052324-25UL; PCK1 (1:1000), Santa Cruz, sc-74825; LEPR (1:500), Novus Bio, NB120-5593; FOXO1 (1:1000), Cell Signaling, 2880 S; CREB1 (1:1000), Cell Signaling, 9197 S; GAPDH (1:4000), Cell Signaling, 2118 S.
QPCR
Total RNA was extracted from tissues and cells using TRI reagent (MRC, TR118, Cincinnati, OH) and reverse transcribed by using Applied Biosystems™ high-capacity reverse transcription kit (Thermo Fisher). Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems) using an ABI Vii7a Real-Time PCR System (Applied Biosystems, Waltham, MA). The sequences of primers used are listed in Supplementary Table 2. mRNA levels were normalized to Rplp0.
RNAseq and data analysis
The liver total RNA from 3 VSG and 3 sham mice was extracted by the Direct-Zol kit (Zymo Research), respectively, and used for the polyA-stranded-RNAseq library preparation. The sequencing was performed by Illumina Hiseq2500 at the City of Hope Integrative Genomics Core. FastQC (version 0.10.1) was used for quality control and STAR (version 2.6.0.a) was used for the alignment to the mouse reference genome (mm10). The read counts were calculated by featureCounts in the Subread package (2.0.1). R (version 4.2.0) and DESeq2 (version 1.37.0) were used for the differential expression analysis. The counts were presented as FPKM (Fragments Per Kilobase Million). The heatmap was drawn by using pheatmap (version 1.0.12). The RNAseq data can be retrieved from the NCBI GEO database (GSE218931).
ChIRP and data analysis
About 250 million primary hepatocytes were isolated from four female 8-week-old WT C57BL/6 mice. These hepatocytes were fixed in 90 mL 3% formaldehyde in 1x DPBS for 10 min. Then 10 mL 1.25 M glycine was added to quench the formaldehyde. The hepatocytes were washed by 1x DPBS, and aliquoted by 60 million cells per tube in ice-cold DPBS and stored at −80 °C. The formaldehyde-fixed hepatocytes were sonicated in 0.5 mL standard ChIRP lysis buffer per tube at the setting of 30-s ON / 30-s OFF for 11 cycles, and then centrifuged at 16,100×g for 10 min, 4 °C. The supernatant was combined and aliquoted to 1 mL/tube. Fourteen probes with antisense to different regions of Gm19619 were divided into two sets (odd- and even-numbered) for the assay, 100 µL Dynabead C1 (Thermo Fisher) per sample was used to pulldown the chromatin fragments according to the JOVE protocol41 and sent for ChIP-seq.
ChIRP-seq was analyzed following the pipeline described by ref. 42. MACS2 was used for peak calling20. ChIPseeker (1.30.3) and ChIPpeakAnno (3.28.1) were used to annotate the peaks and find the overlapping peaks among different samples43,44. The MATCH tool of TRANSFAC (Version 2.0) was used for TFs prediction45. The sequences of ChIRP probes are listed in Supplementary Table 1. The ChIRP-seq data can be retrieved from the NCBI GEO database (GSE218931).
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism (version 9). All data are expressed as mean ± SEM and comprise ≥3 biologically independent replicates if not stated otherwise. All mice experiments were conducted at least twice with 5–11 mice in each group based on sample availability. Statistical significance was analyzed using a two-tailed unpaired Student’s t-test. The threshold of statistical significance was set at P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The uncropped western blot images for Figs. 2c, 3e, 5h, i are in Supplementary Figure 7. The RNAseq and ChIRP-seq data that support the findings of this study have been deposited in the NCBI GEO database (GSE218931, GSE218904, and GSE218905). All other data were available from the corresponding author on reasonable request.
References
Welbourn, R. et al. Bariatric surgery worldwide: baseline demographic description and one-year outcomes from the fourth IFSO Global Registry Report 2018. Obes. Surg. 29, 782–795 (2019).
Arterburn, D. E., Telem, D. A., Kushner, R. F. & Courcoulas, A. P. Benefits and risks of bariatric surgery in adults: a review. JAMA 324, 879–887 (2020).
Ryan, K. K. et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 509, 183–188 (2014).
McGavigan, A. K. et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut 66, 226–234 (2017).
Ding, L. et al. Vertical sleeve gastrectomy activates GPBAR-1/TGR5 to sustain weight loss, improve fatty liver, and remit insulin resistance in mice. Hepatology 64, 760–773 (2016).
Ding, L. et al. Vertical sleeve gastrectomy confers metabolic improvements by reducing intestinal bile acids and lipid absorption in mice. Proc. Natl Acad. Sci. USA 118, e2019388118 (2021).
Du, J. et al. Vertical sleeve gastrectomy reverses diet-induced gene-regulatory changes impacting lipid metabolism. Sci. Rep. 7, 5274 (2017).
Ding, L. et al. Targeting bile acid-activated receptors in bariatric surgery. Handb. Exp. Pharm. 256, 359–378 (2019).
Jin, L. H., Fang, Z. P., Fan, M. J. & Huang, W. D. Bile-ology: from bench to bedside. J. Zhejiang Univ. Sci. B 20, 414–427 (2019).
Dimitriadis, G. K., Randeva, M. S. & Miras, A. D. Potential hormone mechanisms of bariatric surgery. Curr. Obes. Rep. 6, 253–265 (2017).
Holst, J. J. et al. Mechanisms in bariatric surgery: gut hormones, diabetes resolution, and weight loss. Surg. Obes. Relat. Dis. 14, 708–714 (2018).
Sallam, T. et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016).
Hennessy, E. J. et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat. Metab. 1, 98–110 (2019).
Dallner, O. S. et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat. Med. 25, 507–516 (2019).
Lin, M. F., Jungreis, I. & Kellis, M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–282 (2011).
Lizio, M. et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16, 22 (2015).
Scotto-Lavino, E., Du, G. & Frohman, M. A. 5’ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2555–2562 (2006).
Scotto-Lavino, E., Du, G. & Frohman, M. A. 3’ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2742–2745 (2006).
Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Gaspar, J. M. Improved peak-calling with MACS2. Preprint at bioRxiv https://doi.org/10.1101/496521 (2018).
Chen, H. et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell 84, 491–495 (1996).
Oster, M. et al. Hepatic p53 is regulated by transcription factor FOXO1 and acutely controls glycogen homeostasis. J. Biol. Chem. 298, 102287 (2022).
Huynh, F. K. et al. A role for hepatic leptin signaling in lipid metabolism via altered very low density lipoprotein composition and liver lipase activity in mice. Hepatology 57, 543–554 (2013).
Huynh, F. K. et al. Disruption of hepatic leptin signaling protects mice from age- and diet-related glucose intolerance. Diabetes 59, 3032–3040 (2010).
Stefater, M. A. et al. Sleeve gastrectomy induces loss of weight and fat mass in obese rats, but does not affect leptin sensitivity. Gastroenterology 138, 2426–2436 (2010).
Arble, D. M., Schwartz, A. R., Polotsky, V. Y., Sandoval, D. A. & Seeley, R. J. Vertical sleeve gastrectomy improves ventilatory drive through a leptin-dependent mechanism. JCI Insight 4, e124469 (2019).
Huang, X. et al. Hepatic leptin signaling improves hyperglycemia by stimulating MAPK phosphatase-3 protein degradation via STAT3. Cell Mol. Gastroenterol. Hepatol. 14, 983–1001 (2022).
Qin, W. et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 44, 6423–6433 (2016).
Patel, N., Murr, M., Lui, A., Shi, Y. & Cai, J. OR01-6 LncRNA GAS5 directed therapeutic increases insulin receptor expression in adipocytes. J. Endocr. Soc. https://doi.org/10.1210/js.2019-OR01-6 (2019).
Liang, Y. et al. Duodenal long noncoding RNAs are associated with glycemic control after bariatric surgery in high-fat diet-induced diabetic mice. Surg. Obes. Relat. Dis. 13, 1212–1226 (2017).
Castro-Mondragon, J. A. et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–d173 (2022).
Bideyan, L., Nagari, R. & Tontonoz, P. Hepatic transcriptional responses to fasting and feeding. Genes Dev. 35, 635–657 (2021).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e614 (2018).
Zhu, S. Y. et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 34, 1279–1286 (2016).
Ruan, X. et al. In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat. Commun. 11, 45 (2020).
Ruan, X. et al. Identification of human long noncoding RNAs associated with nonalcoholic fatty liver disease and metabolic homeostasis. J. Clin. Invest. 131, e136336 (2021).
M, S. Z., Hartford, C. C. R. & Lal, A. Interrogating lncRNA functions via CRISPR/Cas systems. RNA Biol. 18, 2097–2106 (2021).
Fan, M. et al. Bile acid-mediated activation of brown fat protects from alcohol-induced steatosis and liver injury in mice. Cell Mol. Gastroenterol. Hepatol. 13, 809–826 (2022).
Zhang, E. et al. Intestinal AMPK modulation of microbiota mediates crosstalk with brown fat to control thermogenesis. Nat. Commun. 13, 1135 (2022).
Chu, C., Quinn, J. & Chang, H. Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. https://doi.org/10.3791/3912 (2012).
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Zhu, L. J. et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinforma. 11, 237 (2010).
Matys, V. et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 374–378 (2003).
Acknowledgements
We would like to thank Drs. Arthur Riggs, Rama Natarajan, and other members at AR-DMRI for their discussion and suggestions. This study was supported partially by the George & Irina Schaeffer, the John Hench, and the Ella Fitzgerald Foundations and the National Institutes of Health grants (R01DK124627 to W.H., R01HL145170 and R01GM141096 to Z.B.C., and COH P30CA33572 to COH).
Author information
Authors and Affiliations
Contributions
W.H. conceptualized and designed the experiments and revised the manuscript. Z.F. and M.F. designed the experiments, analyzed data, wrote the manuscript, and performed the bulk of the experiments. L.J., Y.W., J.T., L.D., E.Z., and S.X. helped to collect data. D.Y., X.W., and Z.B.C. analyzed the ChIRP-seq data. Z.F., M.F., Z.B.C., and W.H. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Zhijuan Qiu. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fang, Z., Fan, M., Yuan, D. et al. Downregulation of hepatic lncRNA Gm19619 improves gluconeogenesis and lipogenesis following vertical sleeve gastrectomy in mice. Commun Biol 6, 105 (2023). https://doi.org/10.1038/s42003-023-04483-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-04483-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.