Abstract
To better understand the genetics of hearing loss, we performed a genome-wide association meta-analysis with 125,749 cases and 469,497 controls across five cohorts. We identified 53/c loci affecting hearing loss risk, including common coding variants in COL9A3 and TMPRSS3. Through exome sequencing of 108,415 cases and 329,581 controls, we observed rare coding associations with 11 Mendelian hearing loss genes, including additive effects in known hearing loss genes GJB2 (Gly12fs; odds ratio [OR] = 1.21, P = 4.2 × 10−11) and SLC26A5 (gene burden; OR = 1.96, P = 2.8 × 10−17). We also identified hearing loss associations with rare coding variants in FSCN2 (OR = 1.14, P = 1.9 × 10−15) and KLHDC7B (OR = 2.14, P = 5.2 × 10−30). Our results suggest a shared etiology between Mendelian and common hearing loss in adults. This work illustrates the potential of large-scale exome sequencing to elucidate the genetic architecture of common disorders where both common and rare variation contribute to risk.
Similar content being viewed by others
Introduction
The loss of hearing can have a debilitating impact on quality of life, requiring major adjustments to day-to-day activities. Serious comorbidities are also associated with hearing loss including social isolation, depression, cognitive impairment, and dementia, which further deteriorate quality of life1. Disabling hearing loss is common, with ~466 million people affected worldwide (World Health Organization: https://www.who.int/health-topics/hearing-loss). While existing technologies—such as hearing aids and cochlear implants—can ameliorate hearing loss, their use is limited by barriers to access, including cost, health policies and regulations, and social stigma associated with device use2. Furthermore, while these assistive devices typically provide some benefit, they do not address the chief complaint associated with acquired hearing loss: lack of hearing clarity, particularly in social and work environments (https://www.hearingloss.org/programs-events/patient-focused-drug-development-meeting). The Lancet Commission on dementia prevention, intervention, and care has identified untreated hearing loss in middle age as the top modifiable risk factor for dementia, but it is estimated that 67–86% of adults who may benefit from hearing aids do not use them1,3. These challenges, combined with a lack of therapeutics to stop or slow hearing loss progression, have contributed to its status as a growing global health issue. Novel therapies based on genetic evidence, therefore, will be crucial in addressing this unmet need.
Hearing loss affects individuals of all ages, but its prevalence increases with age. Approximately 1-2/1000 babies are born with hearing loss4. Mutations in over 150 genes (https://hereditaryhearingloss.com) account for over 50% of the cases. While autosomal recessive hearing loss is generally pre-lingual and non-progressive, autosomal dominant forms are mostly post-lingual (including adult onset) and progressive. The prevalence of hearing loss increases to 2.8/1,000 in primary school-age children and 3.5/1000 in adolescents4,5. The National Institute on Deafness and other Communication Disorders calculates that by the age of 45 ~2% of individuals have a disabling hearing loss, and this number increases to 50% in individuals over the age of 75. This increase in prevalence with age reflects a combination of late-onset hearing loss mutations, the cumulative effects of environmental factors such as exposure to noise and ototoxic drugs and, in aging individuals, the degenerative effects of age on the cochlea. These genetic and environmental insults primarily damage the structures of the inner ear, resulting in sensorineural hearing loss6,7.
Heritability estimates for age-related hearing loss range as high as 36 and 70%8,9,10,11,12 suggesting that genetics, along with environmental factors, play a major role in determining an individual’s risk for developing hearing loss. Genome-wide association studies (GWAS) of hearing loss in adults have identified 61 common variant loci associated with the trait in Europeans13,14,15,16,17,18,19,20. Three of these studies18,19,20 have included genotyping and imputed data from UK Biobank as well. While the majority of these studies have established a common variant contribution to adult hearing loss, few reports have addressed the contibution of rare and low-frequency variation21.
Recently, Ivarsdottir et al.18 published association results with hearing loss on ~50,000 Icelandic individuals with whole-genome sequence and ~50,000 individuals from UKB with exome sequence data, with imputation of larger samples into these variant sets. We have now expanded the rare variant analysis to exome-sequences from ~294,710 individuals in UKB and ~143,286 individuals from three other datasets. Here, we report findings from genome- and exome-wide association meta-analyses including 125,749 cases and 469,497 controls. Our analyses have identified 15 susceptibility loci that were previously not associated with hearing loss, to the best of our knowledge, and 15 rare variant associations that provide important insights into the biology of hearing loss in adults.
Results
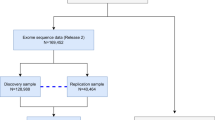
To study common variants, we performed association meta-analyses using genotyping and imputation across five cohorts: the Geisinger DiscovEHR study (GHS), the Malmö Diet and Cancer study from Malmö, Sweden (MALMO), the Mount Sinai’s BioMe Personalized Medicine Cohort from Mount Sinai Health System, New York (SINAI), UK Biobank (UKB) and an additional study from Finland, FinnGen, for a total of 125,749 cases and 469,497 controls. To study rare and ultra-rare variants, we also generated exome sequence data and performed combined GWAS and exome-wide association study (ExWAS) on a subset of 108,415 cases and 329,581 controls across GHS, MALMO, SINAI, and UKB. Phenotypes were derived from ICD-10 diagnosis codes in GHS, MALMO, SINAI, and FinnGen, and combined self-report and ICD-10 codes in UKB (see Methods and Supplementary Data 1 for details). Our genome-wide association analyses included 15,881,489 variants with frequency > 0.1% that were genotyped or imputed with r2 > 0.3 in at least one study, and 2,923,124 coding or essential splice site variants with minor allele count at least five from exome sequencing (111,588 of which overlapped with the imputed).
Common variant associations
We identified 53 independent loci harboring genome-wide significant (P < 5 × 10−8) common (MAF ≥ 0.01) variants associated with hearing loss (Fig. 1a, Supplementary Data 2, Supplementary Fig. 1), 38 of which are shared with the previously reported 61 hearing loss-associated loci. We observed a genomic control lambda_GC = 1.36 but an LD score regression intercept 1.054 (standard error 0.008), indicating that inflation was largely due to polygenic signal for the hearing loss phenotype.
Among the lead variants at the 15 loci uncovered in this analysis (Fig. 1a, Table 1, Supplementary Fig. 2) is rs117887149 that maps close to GJB2 (Supplementary Fig. 2g), which is a predominant cause of congenital hearing loss. While the majority of lead variants in these loci lay in intergenic or downstream/upstream regions of genes, at two loci they were within the introns of the following genes: KCTD10, a member of the potassium channel tetramerization domain family that is implicated in cardiac development22,23 and MAP7D2, an axonal cargo transport protein predominantly expressed in the brain24.
Top variants at two other loci were missense changes and thus also implicate specific genes: in COL9A3 (Arg103Trp; MAF = 0.07) and TMPRSS3 (Ala90Thr; MAF = 0.06). Mutations in COL9A3, which is highly expressed in the ear, have been implicated in the autosomal recessive Stickler syndrome, in which hearing loss is prominent25, and tentatively in non-syndromic hearing loss26,27. High-throughput mouse knockout characterization from the International Mouse Phenotyping Consortium (IMPC; https://www.mousephenotype.org) indicates that Col9a3-null mice also have hearing loss28. TMPRSS3 is a type II membrane serine protease that localizes to the endoplasmic reticulum and plasma membranes, and is expressed in hair cells and supporting cells in the organ of Corti, the spiral ganglion, and the stria vascularis in the ear29,30,31. Mutations in TMPRSS3 cause congenital and childhood-onset autosomal recessive hearing loss32 but there is also evidence for hearing deficits in heterozygous carriers18.
The number of associated variants at the 50 autosomal loci ranged from eight to 1326. In order to assess the presence of multiple independent causal variants at each locus, we ran conditional analyses using GCTA-COJO33, which indicated the presence of secondary association signals (joint P < 10−5) at eight loci (Supplementary Data 3). We also ran FINEMAP Bayesian causal variant inference34 to prioritize associated variants as credibly causal at thirty loci that were genome-wide significant in RGC data (excluding FinnGen). The COJO and FINEMAP methods disagreed on the presence of single vs. multiple association signals within five loci, three of which showed subthreshold (10−5 < P < 10−4) secondary associations in COJO. FINEMAP prioritized ten or fewer variants in top causal variant 95% credible sets for the top causal variant at eleven loci, including single putatively causal variants at four loci (Supplementary Data 3): missense variants in CDH23 (Ala371Thr; rs143282422) and KLHDC7B (Val504Met; rs36062310), and intronic variants in CTBP2 (rs10901863) and PAFAH1B1 (rs12938775).
Colocalization with GTEx eQTLs identifies candidate genes driving GWAS signals
To identify genes for which expression regulation might drive the observed association signals, we tested for colocalization of our hearing loss-associated loci with expression quantitative trait loci (eQTL) data for 48 tissues from the Genotype-Tissue Expression (GTEx; https://www.gtexportal.org) project using coloc235. Across all GTEx eQTL tissues tested, we identified 19 genes mapping to 15 loci with evidence (posterior probability of colocalization, PPH4 ≥ 0.5) for colocalization between the hearing loss association and an eQTL signal, in at least one tissue (Supplementary Data 4). Only two of the 15 loci with GWAS-eQTL overlap had multiple eQTL signals for more than one gene (NUCKS1 and RAB29 in locus 1-4; ACADVL, DLG4, CTDNEP1, and CLDN7 in locus 17-2) making it difficult to prioritize causal genes at these loci based on eQTL data. Since GTEx did not include tissues from the ear, we used single-cell RNA sequencing data that were generated in-house from mouse cochleae to check the expression of the 19 genes in the ear (Supplementary Data 5). Thirteen of the 19 genes showed evidence of expression across 26 inner-ear cell types and, of these, 12 were expressed in the hair cells. While the majority of genes showed broad expression across the 26 cell types, we noted a subset that were specific to only a few, including CRIP3 in inner and outer hair cells, and TCF19 in neurons and immune cells (Supplementary Data 4 & 5).
Rare-variant association analysis identifies large-effect hearing loss variants in known hearing loss genes
We identified significant (P < 5 × 10−8) rare variant (MAF < 0.01) associations in 25 genes, 15 of which had nonsynonymous variant or gene burden associations (Fig. 1b, Table 2, Supplementary Data 6 & 7). Five of the 15 genes also had significant common variant associations within 1 Mb (KLHDC7B, SYNJ2, GJB2, EYA4, and CDH23). After conditioning on independent common variants at each locus (Supplementary Data 6, 7 & 8), the rare variant and gene burdens remained associated with hearing loss (maximum conditional P ≤ 3 × 10−5) except for the CDH23 Asn1103Ser association (conditional P = 0.79). Of note, the lead common variant in CDH23 is also a missense (Ala371Thr) variant (MAF = 0.01) and is pinpointed by FINEMAP as the only causal variant at that locus with high confidence. Overall, our conditional analyses suggest that rare variant association signals are usually independent of nearby common variant associations.
Associations with Mendelian hearing loss genes
Of the 14 genes with independent, rare nonsynonymous and/or burden associations, 11 were previously identified as causes of Mendelian forms of hearing loss. These include associations with two genes (GJB2 and SLC26A5) that cause recessive hearing loss, burden associations in seven genes (MYO6, COCH, TECTA, SIX1, CEACAM16, POU4F3, and EYA4) and single-variant associations in two genes (TBC1D24 and COL11A2) that cause autosomal dominant hearing loss (reviewed in Shearer et al.36). In MYO6 and COCH, we also observed genome-wide significant single-variant associations of large-effect size (MYO6 His246Arg, OR = 30.7; COCH Cys542Phe, OR = 81.4; Supplementary Data 6). Both variants have previously been characterized as pathogenic in family-based genetic analyses37,38. Only a minority of variants included in burden tests have been classified as pathogenic by ClinVar (Supplementary Data 9) suggesting that our analysis has detected additional, risk-associated variants with variable penetrance in Mendelian hearing loss genes. We also identified single-variant associations in two genes that cause autosomal dominant hearing loss: TBC1D24 Asn307Ser, also recently reported by Ivarsdottir et al.18, was implicated as pathogenic in two unrelated families39, and COL11A2 Phe80Ser, which has not yet been classified as pathogenic but is predicted deleterious, lies in a domain (Laminin G-like/NC4) that harbors other mutations causing non-syndromic hearing loss40.
We performed our analysis under an additive model, which assumes risk effects in heterozygous carriers as well as homozygotes; therefore, it is not surprising to see that the majority of Mendelian genes (9/11) identified can cause hearing loss in heterozygote carriers. However, we also detect associations in two genes that have previously been implicated in recessive hearing loss: GJB2 Gly12fs (OR = 1.21; P = 4 × 10−11), and SLC26A5 Leu46Pro (OR = 1.3; P = 3 × 10−14) as well as the burden of SLC26A5 predicted loss-of-function (pLOF) and strict deleterious missense variants (excluding Leu46Pro) (OR = 1.96; P = 3 × 10−17) (Fig. 2). We also observed a suggestive association with GJB2 Leu90Pro (OR = 1.51, P = 4.4 × 10−5), another known pathogenic variant in recessive hearing loss. The associations persisted after excluding homozygous carriers and compound heterozygous carriers of rare, coding variants in these genes (Supplementary Data 10), suggesting a previously unappreciated increase in risk for hearing loss in heterozygous carriers of loss-of-functionGJB2, and missense and pLOF SLC26A5 variants.
Rare-variant associations in KLHDC7B, FSCN2, and SYNJ2
The most significant rare coding association in our analysis was the aggregate of 68 pLOF variants (43 frameshift, 23 stop-gain and 2 stop-loss, (Supplementary Data 9)) in KLHDC7B (Kelch-like domain containing 7B), with an approximately two-fold increase in risk for hearing loss (OR = 2.14, P = 5 × 10−30). The main contributors to the gene burden were two frameshift variants, Gly302fs and Lys181fs, that are predicted to truncate the protein near the start of the Kelch domains. These variants were also significantly associated in single-variant tests (Fig. 3), and Gly302fs was recently reported in an analysis that included UKB18. The association with pLOF variants remained significant after repeating the pLOF burden test conditioning on Gly302fs and Lys181fs (P = 8 × 10−8; Supplementary Data 7). In addition, we observed a common (MAF = 0.04) missense variant (Val504Met) within the last Kelch domain of KLHDC7B associated with increased risk for hearing loss (OR = 1.14, P = 4 × 10−26). This variant was also prioritized as the sole causal variant at this locus by FINEMAP (Supplementary Data 3).
We also identified rare coding associations in fascin actin-bundling protein 2 (FSCN2) and synaptojanin 2 (SYNJ2) with increased risk for hearing loss, both of which were recently observed in UKB18 (Fig. 4). In FSCN2, an actin cross-linking protein41,42,43, an aggregate of pLOF and deleterious missense variants was associated with hearing loss, with majority of the carriers in the burden having the His138Tyr variant. Conditioning on His138Tyr attenuated the significance of the burden test (P = 3 × 10−6 after conditioning on His138Tyr; Supplementary Data 7) but did not eliminate the signal, suggesting that other variants in FSCN2 may increase the risk for hearing loss. Mice homozygous for loss-of-function mutations in Fscn2 present progressive hearing loss starting at 3 weeks and near deafness by 24 weeks due to degeneration of the outer hair cells in the cochlea43. In SYNJ2, we identified an association with a missense (Thr656Met)18 variant that lies in the catalytic domain of this inositol polyphosphate 5-phosphatase44. Mice harboring homozygous mutations in Synj2 that are predicted to reduce protein levels or the 5-phosphatase catalytic activity show progressive high-frequency hearing loss and a degeneration of hair cells that is most profound in the outer hair cells45,46.
GWAS/ExWAS supports a highly polygenic architecture of adult hearing loss
Given that this is the largest sequencing study to date of adult hearing loss, and given the presence of Mendelian hearing loss genes among our common and rare-variant associations, we sought to explore the distribution of effect sizes across allele frequencies (Fig. 5). Hearing loss-associated variants span the frequency spectrum and, while we observe a few rare variants of large effect (e.g. COCH Cys542Phe and MYO6 His246Arg), we do not observe any common variants of large effect. We further estimated phenotypic variance explained by the genetic data, or heritability h2Tot, using LD score regression (LDSC) partitioning into functional categories and stratifying by minor allele frequency47,48. We estimated a total heritability (h2Tot) of 0.089, with contributions from common variation (h2CV, MAF > 0.05) and low-frequency variation (h2LFV, 0.001 < MAF ≤ 0.05) of 0.074 and 0.015, respectively (Supplementary Fig. 3, Supplementary Data 11). These results indicate that, while the bulk of SNP heritability is derived from common variation, low-frequency variation contributes 16.8% of total SNP heritability.
Plotted are odds ratio estimates (on log scale) and minor allele frequencies of genome-wide significant variants and gene burdens (Supplementary Data 2, 6 and 7). 50% and 80% power curves for the present study are plotted as dotted lines. Notably, almost all points (particularly low-frequency and common variants, MAF > 0.0005) lie very close to the dotted power curves.
Discussion
We performed combined GWAS and ExWAS using exome-sequencing data and identified 53 independent associations, of which 15 have not previously been associated with hearing loss, to the best of our knowledge. The 15 associations included a missense lead variant in TMPRSS3, a known cause of Mendelian hearing loss, adding to the tally of Mendelian deafness genes (EYA4, CDH23, TRIOBP) showing common coding-variant associations with hearing loss in adult humans. We also identified coding variant/gene burden associations with 15 genes through exome sequencing. We estimated that low-frequency variation contributes a non-negligible portion (16.8%) of SNP heritability for adult hearing loss, consistent with a highly polygenic genetic architecture with rare, low-frequency and common genetic variation for adult hearing loss, and where variants of large effect would be subject to purifying selection.
Notably, the majority of genes implicated by rare-variant and burden associations are already known to cause Mendelian forms of hearing loss; however, the odds ratios for these associations are wide-ranging. At the lower end of the effect spectrum are single-variant associations in GJB2 and SLC26A5, with ORs close to typical for GWAS findings (1.2-1.3), followed by COL11A2 (2.2-7) and, at the high end of the distribution, COCH and MYO6 (31-81). COCH Cys542Phe and MYO6 His246Arg are known pathogenic variants, and consistent with this, we observe almost all carriers presenting hearing loss. The two control carriers for each variant (Supplementary Data 6) could be explained by imperfect ascertainment of the phenotype or, as these mutations cause late-onset and progressive hearing loss, could also reflect a difference in expressivity. While some dilution of the effect sizes may be expected when working with self-reported as opposed to objective measures of hearing loss, the broad range in ORs that we observe suggests that for several of these Mendelian hearing loss genes we are identifying carriers with incompletely penetrant, risk-increasing variants. These results are consistent with the hypothesis that there is a continuum between Mendelian and common forms of hearing loss with the same genes harboring mutations causal for the former, and risk-increasing for the latter. Furthermore, compared to epidemiological risk factors for common hearing loss, including noise exposure (OR ~ 1.5–349), odds ratios of low-frequency and rare genetic factors may be large enough to provide mechanistic insights and to have implications for precision medicine.
Ultimately, to understand the biology of hearing loss and develop testable hypotheses from our association results, we need to identify the genes driving the observed associations at each locus. For common variants, we note the high resolution of FINEMAP to prioritize causal variants in several loci, including pinpointing single variants in four genes, three of which are related to hearing loss (CDH23), hearing function (CTBP2), or have significant rare variant associations with hearing loss (KLHDC7B). While FINEMAP helps prioritize variants, the gene that is impacted by those variants is not necessarily obvious, so we looked for colocalization with GTEx eQTL data and identified 19 genes whose expression may influence hearing loss risk (Supplementary Data 4). Analysis of single-cell expression data from mouse ears showed that 13 of the 19 genes are expressed in ear tissues. Of note, among these are LMO7 and SPTBN1 (BetaII-spectrin). Both genes code for components of the cuticular plate in the hair cells of the ear and knockout mice for either gene develop hearing loss50,51. LMO7 colocalizes with eQTLs for the gene in GTEx skeletal muscle and SPTBN1 in thyroid tissue, such that decreased expression of these genes is correlated with increased risk for hearing loss in humans.
One of the most compelling ways in which human genetics can establish a role for a gene in disease is when a diverse set of rare nonsynonymous or loss-of-function variants in the same gene show consistent association with disease. Coding variant and gene burden associations that we identified included associations with known pathogenic (e.g. in MYO6 and COCH) and uncharacterized variants (COL11A2 Phe80Ser), in genes that can cause autosomal dominant Mendelian hearing loss. We also identified variants and genes that cause recessive hearing loss (GJB2 Gly12fs and SLC26A5 pLOF+strict deleterious missense mutations), associated with increased risk (OR~1.2–1.3) for hearing loss in heterozygous carriers. Missense mutations in GJB2 can cause dominant, syndromic hearing loss52 but loss-of-function mutations, including Gly12fs, cause recessive non-syndromic hearing loss53,54,55. Consistent with our findings, studies have detected hearing deficiency at high frequencies and a possibly more prominent effect in female adult heterozygous carriers of Gly12fs55,56,57. Mutations in SLC26A5 cause recessive hearing loss in humans58 and mice homozygous for Slc26a5 null mutations develop hearing loss as early as 5–7 weeks of age59. Slc26a5 heterozygous null mice have a hearing deficiency that is intermediate between the controls and knockouts. While we cannot rule out the possibility of compound heterozygosity with variation in regulatory/promoter regions in heterozygous carriers, our results offer the possibility that heterozygous carriers of loss-of-function variants in GJB2 and SLC26A5 may also have increased risk for adult-onset hearing loss.
Three of the 15 coding and burden associations were in genes that have not previously been implicated in Mendelian hearing loss in humans: KLHDC7B, FSCN2, and SYNJ2. In KLHDC7B, we confirmed a previously reported association of the common Val504Met and a rare frameshift variant with increased risk for hearing loss18,20, and identified associations of increased risk with a burden of additional pLOF variants. While the biological effect of the missense is difficult to interpret, the association of putative LOFs with increased risk for hearing loss suggests that loss of KLHDC7B function is deleterious for hearing function. Based on the smaller effect of Val504Met (OR = 1.14) compared to the pLOF burden (OR = 2.14), we would hypothesize that this more common missense (MAF = 0.04) is a hypomorph. KLHDC7B (Kelch-like domain containing 7B) is a relatively understudied gene; it has been characterized as a 594-amino-acid protein containing a Kelch domain that is hypermethylated and upregulated in breast cancer cell lines and may influence cell proliferation in MCF-7 cells lines60,61. In the mouse ear, RNAseq expression profiling62 and our real-time PCR data (Supplementary Fig. 4) show Klhdc7B expression in the cochlea with enrichment in the outer hair cells63. Consistent with the hypothesis from our genetic findings that loss of KLHDC7B function increases risk for hearing loss, initial characterization of Klhdc7B null mice by IMPC showed that homozygous carriers have hearing loss28. Mouse models Fscn2 and Synj2 also develop hearing loss43,45,46. Based on our association results, it would also be interesting to test heterozygous null Klhdc7b, Fscn2, and Synj2 animals for increased susceptibility to hearing loss with age or environmental insults such as noise exposure.
We recognize that our study has several limitations. Our phenotype includes (in UKB) self-reported hearing loss among adults, which is likely to be a heterogeneous mix of early-onset, late-onset, age-related, as well as hearing loss due to environmental insults. In general, greater phenotype precision, including environmental exposure measures, should help future genetic analyses of adult hearing loss. We note that our colocalization analyses utilized GTEx eQTL data across tissues not including ear expression data. Given the high degree of sharing of genetic regulation of expression across tissues64,65, the results are likely to point to the causal genes in many instances. eQTL data for ear cell types should help with the interpretation of genetic analyses of adult hearing loss.
In summary, this work contributes to connecting the two ends of the genetic architecture of hearing loss by detecting a common signal in genes known to cause hearing loss in Mendelian fashion and by detecting an additive signal (i.e. increased risk in heterozygous carriers) in genes known to cause autosomal recessive hearing loss. This latter finding also connects young- and adult-onset hearing loss in a single phenotypic spectrum with complex genetic underpinnings, including contributions from rare and common variation.
Methods
Participating cohorts and phenotype data
We performed meta-analysis for hearing loss on a total of 125,749 cases and 469,497 controls of European ancestry from the following cohorts: United Kingdom Biobank (UKB)66, the MyCode Community Health Initiative cohort from Geisinger Health System (GHS)67, the Mount Sinai BioMe cohort (SINAI) (https://icahn.mssm.edu/research/ipm/programs/biome-biobank/pioneering), the Malmö Diet and Cancer study (MALMO)68, and FinnGen R3 (https://www.finngen.fi/). Hearing loss case-control status in GHS, MALMO, and SINAI was defined using ICD-10 code diagnoses from the EHR. Cases were individuals with ICD10 H90.3-H90.8, H91.1or H91.9 diagnoses. Controls were individuals who did not meet the case criteria and did not have a diagnosis for ICD-10 Q16 (congenital malformations of ear causing hearing impairment) or ICD-10 H93.1 (tinnitus). In UKB the phenotype was defined using ICD-10 codes as above or by self-reported hearing loss or complete deafness on the touchscreen questionnaire (Field IDs: 2247 and 2257). Controls in UKB were individuals who did not have an ICD-10 diagnosis for hearing loss or tinnitus and did not self-report hearing loss, deafness or tinnitus (Field IDs: 4803 and non-cancer illness code 1597). The FinnGen analysis used was finngen_r3_H8_CONSENHEARINGLOSS, conductive or sensorineural hearing loss defined by ICD10 codes H90[.0-8], versus controls excluding other ear disorders (H91-H95). Further details are given in the Supplementary Information.
Ethical approval and informed consent
All participants provided informed consent, and studies were approved by the individual Institutional Review Boards (IRBs) at the respective institutions. UK Biobank has approval from the North West Multi-Centre Research Ethics Committee (MREC; ref: 11/NW/0382), which covers the UK. It also sought the approval in England and Wales from the Patient Information Advisory Group (PIAG) for gaining access to information that would allow it to invite people to participate. The DiscovEHR study was approved by the Geisinger Health System Institutional Review Board. The BioMe Biobank is an ongoing research biorepository approved by the Icahn School of Medicine at Mount Sinai’s IRB. The Ethical Committee at Lund University approved the Malmo Diet and Cancer Study (LU 51-90). The Finngen Biobank was approved by the Coordinating Ethics Committee of the Helsinki and Uusimaa Hospital District.
Genetic data and association analyses
High-coverage whole-exome sequencing was performed at the Regeneron Genetics Center as previously described69,70. For SINAI and MALMO, DNA from participants was genotyped on the Global Screening Array (GSA), and for GHS genotyping was done on either the Illumina OmniExpress Exome (OMNI) or GSA array; the datasets (stratified by array for GHS) were imputed to the TOPMed (GHS) or HRC (MALMO and SINAI) reference panels using the University of Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html) or the TOPMed Imputation Server (https://imputation.biodatacatalyst.nhlbi.nih.gov/). Additional details are given in the Supplementary Information. We tested for association with hearing loss genetic variants or their gene burdens using REGENIE v1.0.4371. Analyses were adjusted for age, age2, sex, an age-by-sex interaction term, experimental batch-related covariates, and genetic principal components. Cohort-specific statistical analysis details are provided in the Supplementary Information. Results across cohorts were pooled using inverse-variance weighted meta-analysis.
Fine mapping and follow-on genetic analyses
We defined genome-wide significant loci in our analysis by linkage disequilibrium (r2 > 0.1) with lead variants. We defined previously associated loci by their index variants reported in previous hearing loss GWAS13,14,15,16,17,19,20,72,73,74,75, and excluded 1 Mb regions surrounding them in the identification of previously unreported, to the best of our knowledge, loci in our analysis. LD score (LDSC) regression76 was used to assess inflation (LDSC intercept) accounting for polygenic signal. Power calculations determined genotype relative risks (GRRs) providing 80 and 50 percent power given specified risk allele frequencies (RAFs, from 10−6 to 1) and the numbers of cases and controls in our meta-analysis. Fine mapping analyses included forward stepwise conditional analyses carried out in every locus with GCTA-COJO using a UK Biobank subsample LD reference panel, with independent associations determined using a joint P-value threshold of 1 × 10−5 and r2 cutoff of 0.9, and FINEMAP34 Bayesian causal variant inference, using LD from available individual level data.
For genes whose cis regions overlapped genome-wide significant hearing loss loci, coloc235 was used to assess evidence for colocalization between our hearing loss GWAS and GTEx (release v8) cis-eQTL data derived from 48 tissues (https://www.gtexportal.org/home/), using GWAS and eQTL summary statistics for all common (MAF ≥ 0.01) variants within each gene’s cis-region. Genes with posterior probability of colocalization H4 ≥ 0.5 were determined as having evidence for colocalization, and were visually inspected in eQTL+GWAS regional association plots77.
Conditional analyses were performed for each cohort using REGENIE’s Firth-corrected logistic regression and the resulting summary statistics were meta-analyzed as described above. Rare-variant association analyses conditional on the common variant signal were carried out for four loci with both common (MAF ≥ 0.01) and single rare variant (MAF < 0.01) genome-wide significant signals, by including as covariates the dosages of variants identified in fine mapping analyses. Burden analyses conditional on rare variants were carried out for five genes with significant single rare variant and burden associations.
Assessment of heterozygous effects used association analyses excluding homozygotes as well as individuals carrying pairs of exome sequenced variants (MAF < 0.02 and MAC > 1) that were called as compound heterozygous mutations (CHMs) or potential CHMs (i.e. unknown phase). CHMs were called by SHAPEIT4 (https://github.com/odelaneau/shapeit4) phasing of merged genotype and exome data with scaffolds based on inferred close relatives78,79.
Heritability derived from variants partitioned into seven functional categories (coding-synonymous, coding-nonsynonymous, 5-prime-UTR, 3-prime-UTR, splice site, intronic, intergenic) with each category further stratified into a low frequency (0.001 < MAF ≤ 0.05) and common (MAF > 0.05) minor allele frequency bin as in stratified LD score regression was estimated using LD score regression (LDSC) of hearing loss association statistics on LD scores. LD scores were generated from a reference panel of N = 10,000 random UK Biobank European-ancestry samples’ merged imputed and exome data.
Single-cell RNA sequencing and analysis
All protocols were approved by the Institutional Animal Care and Use Committee in accordance with the Regeneron’s Institutional Animal Care and Use Committee (IACUC). Cochlea and utricles from C57BL/6 mice at post-natal day 7 were micro-dissected and dissociated. Suspensions of 200 cells/μL were subjected to Chromium Single Cell (10x Genomics) library preparation and were sequenced on Illumina NextSeq 500. Cell Ranger Single-Cell Software Suite (10x Genomics, v2.0.0) was used to perform sample de-multiplexing, alignment to MM10 Genome assembly with UCSC gene models, filtering, and UMI counting. PCA, UMAP and clustering analyses used Seurat V3.2 (https://github.com/satijalab/seurat)80. Cluster marker genes as well as canonical cell type-specific genes were used to manually label the cell type for each cluster.
Statistics and reproducibility
For genome-wide association meta-analysis, the statistical threshold of P < 5 × 10−8 was considered statistically significant. For simplicity and reproducibility, meta-analysis was performed to combine statistical results across cohorts rather than a multi-stage design or requiring nominal significance in multiple cohorts; sample sizes are reported above and in Supplementary Data 1. In conditional analyses of common variants within loci, P < 5 × 10−8 was considered as a threshold for reporting independent associations within loci. Rare-variant associations were reported as independent of common variants in the same locus when they were P < 5 × 10−3, as determined by Bonferroni correction for 10 conditional tests performed. We describe burden results conditional on individual variants within the aggregate burden counts regardless of significance, in order to determine if any variants were driving the burden signal. Further details are given above and in the Supplementary Note.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All whole-exome sequencing, genotyping chip, and imputed sequence for UKB described in this report are publicly available to registered researchers via the UK Biobank data access protocol. Additional information about registration for access to the data is available at http://www.ukbiobank.ac.uk/register-apply/. Further information about the whole-exome sequence is available at http://www.ukbiobank.ac.uk/wp-content/uploads/2019/03/Access_064-UK-Biobank-50k-Exome-Release-FAQ-v3.pdf Detailed information about the chip and imputed sequence is available at: http://www.ukbiobank.ac.uk/wp-content/uploads/2018/03/UKB-Genotyping-and-Imputation-Data-Release-FAQ-v3-2-1.pdf. Geisinger DiscovEHR, Malmo Diet and Cancer study and Mt. Sinai Biome exome-sequencing and genotyping data can be made available to qualified, academic, non-commercial researchers upon request via a Data Transfer Agreement with the respective institutions. Summary statistics for FinnGen r3 can be downloaded from https://www.finngen.fi/en/access_results. Regeneron materials (RNA sequencing data) described in this manuscript are available to qualified, academic, non-commercial researchers upon request through Regeneron portal (https://regeneron.envisionpharma.com/vt_regeneron/) after signing Material/Data Transfer Agreements with Regeneron. Regeneron may deny any request for RNASeq data made by or on behalf of a recipient outside of the academic community; for any use other than to replicate or extend the results described in this publication, including any commercial use or use sponsored by a commercial entity.
References
Cunningham, L. L. & Tucci, D. L. Hearing loss in adults. N. Engl. J. Med. 377, 2465–2473 (2017).
National Academies of Sciences, Engineering, and Medicine, Health and Medicine Division, Board on Health Sciences Policy & Committee on Accessible and Affordable Hearing Health Care for Adults. Hearing Health Care for Adults: Priorities for Improving Access and Affordability (National Academies Press, 2016).
Livingston, G. et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet 396, 413–446 (2020).
Morton, C. C. & Nance, W. E. Newborn hearing screening–a silent revolution. N. Engl. J. Med. 354, 2151–2164 (2006).
Korver, A. M. H. et al. Congenital hearing loss. Nat. Rev. Dis. Primers 3, 16094 (2017).
Liberman, M. C. Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Res. 6, 927 (2017).
Wu, P.-Z., O’Malley, J. T., de Gruttola, V. & Liberman, M. C. Age-related hearing loss is dominated by damage to inner ear sensory cells, not the cellular battery that powers them. J. Neurosci. 40, 6357–6366 (2020).
Karlsson, K. K., Harris, J. R. & Svartengren, M. Description and primary results from an audiometric study of male twins. Ear Hear 18, 114–120 (1997).
Christensen, K., Frederiksen, H. & Hoffman, H. J. Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J. Am. Geriatr. Soc. 49, 1512–1517 (2001).
Kvestad, E., Czajkowski, N., Krog, N. H., Engdahl, B. & Tambs, K. Heritability of hearing loss. Epidemiology 23, 328–331 (2012).
Hendrickx, J.-J. et al. Familial aggregation of pure tone hearing thresholds in an aging European population. Otol. Neurotol 34, 838–844 (2013).
Viljanen, A. et al. Genetic and environmental influences on hearing in older women. J. Gerontol. A: Biol. Sci. Med. Sci. 62, 447–452 (2007).
Friedman, R. A. et al. GRM7 variants confer susceptibility to age-related hearing impairment. Hum. Mol. Genet. 18, 785–796 (2009).
Hoffmann, T. J. et al. A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet 12, e1006371 (2016).
Nagtegaal, A. P. et al. Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Sci. Rep. 9, 15192 (2019).
Fransen, E. et al. Genome-wide association analysis demonstrates the highly polygenic character of age-related hearing impairment. Eur. J. Hum. Genet. 23, 110–115 (2015).
Van Laer, L. et al. A genome-wide association study for age-related hearing impairment in the Saami. Eur. J. Hum. Genet. 18, 685–693 (2010).
Ivarsdottir, E. V. et al. The genetic architecture of age-related hearing impairment revealed by genome-wide association analysis. Commun Biol 4, 706 (2021).
Kalra, G. et al. Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PLoS Genet. e1009025. https://doi.org/10.1371/journal.pgen.1009025 (2019).
Wells, H. R. R. et al. GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK Biobank. Am. J. Hum. Genet. 105, 788–802 (2019).
Lewis, M. A. et al. Whole exome sequencing in adult-onset hearing loss reveals a high load of predicted pathogenic variants in known deafness-associated genes and identifies new candidate genes. BMC Med. Genomics 11, 77 (2018).
Tong, X. et al. Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nat. Commun. 5, 3153 (2014).
Ren, K. et al. KCTD10 is involved in the cardiovascular system and Notch signaling during early embryonic development. PLoS ONE 9, e112275 (2014).
Pan, X. et al. MAP7D2 localizes to the proximal axon and locally promotes kinesin-1-mediated cargo transport into the axon. Cell Rep. 26, 1988–1999.e6 (2019).
Hanson-Kahn, A. et al. Autosomal recessive Stickler syndrome resulting from a COL9A3 mutation. Am. J. Med. Genet. A 176, 2887–2891 (2018).
Asamura, K., Abe, S., Fukuoka, H., Nakamura, Y. & Usami, S.-I. Mutation analysis of COL9A3, a gene highly expressed in the cochlea, in hearing loss patients. Auris Nasus Larynx 32, 113–117 (2005).
Wonkam, A., Manyisa, N., Bope, C. D., Dandara, C. & Chimusa, E. R. Whole exome sequencing reveals pathogenic variants in MYO3A, MYO15A and COL9A3, and differential frequencies in ancestral alleles in hearing impairment genes among individuals from Cameroon. Hum. Mol. Genet. https://doi.org/10.1093/hmg/ddaa225 (2020).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Guipponi, M. et al. An integrated genetic and functional analysis of the role of type II transmembrane serine proteases (TMPRSSs) in hearing loss. Hum. Mutat. 29, 130–141 (2008).
Fasquelle, L. et al. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J. Biol. Chem. 286, 17383–17397 (2011).
Tang, P.-C. et al. Defective Tmprss3-associated hair cell degeneration in inner ear organoids. Stem Cell Rep. 13, 147–162 (2019).
Scott, H. S. et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 27, 59–63 (2001).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, S1–S3 (2012).
Benner, C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016).
Dobbyn, A. et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 102, 1169–1184 (2018).
Shearer, A. E. et al. (eds) In GeneReviews® [Internet] (University of Washington, Seattle, 1999).
Mohiddin, S. A. et al. Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6). J. Med. Genet. 41, 309–314 (2004).
Street, V. A. et al. A novel DFNA9 mutation in the vWFA2 domain of COCH alters a conserved cysteine residue and intrachain disulfide bond formation resulting in progressive hearing loss and site-specific vestibular and central oculomotor dysfunction. Am. J. Med. Genet. A 139A, 86–95 (2005).
Parzefall, T. et al. A novel variant in the TBC1D24 lipid-binding pocket causes autosomal dominant hearing loss: evidence for a genotype-phenotype correlation. Front. Cell. Neurosci. 14, 585669 (2020).
Chakchouk, I. et al. Novel mutations confirm that COL11A2 is responsible for autosomal recessive non-syndromic hearing loss DFNB53. Mol. Genet. Genomics 290, 1327–1334 (2015).
Hashimoto, Y., Kim, D. J. & Adams, J. C. The roles of fascins in health and disease. J. Pathol. 224, 289–300 (2011).
Perrin, B. J. et al. β-Actin and fascin-2 cooperate to maintain stereocilia length. J. Neurosci. 33, 8114–8121 (2013).
Liu, X. et al. Null mutation of the fascin2 gene by TALEN leading to progressive hearing loss and retinal degeneration in C57BL/6J Mice. G3 8, 3221–3230 (2018).
Rusk, N. et al. Synaptojanin 2 functions at an early step of clathrin-mediated endocytosis. Curr. Biol. 13, 659–663 (2003).
Manji, S. S. M. et al. A mutation in synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain Mozart. PLoS ONE 6, e17607 (2011).
Martelletti, E. et al. Synaptojanin2 mutation causes progressive high-frequency hearing loss in mice. Front. Cell. Neurosci. 14, 561857 (2020).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Gazal, S. et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 50, 1600–1607 (2018).
Gong, R. et al. Hearing loss prevalence and risk factors among older adults in China. Int. J. Audiol. 57, 354–359 (2018).
Du, T.-T. et al. LMO7 deficiency reveals the significance of the cuticular plate for hearing function. Nat. Commun. 10, 1117 (2019).
Liu, Y. et al. Critical role of spectrin in hearing development and deafness. Sci Adv. 5, eaav7803 (2019).
Lee, J. R. & White, T. W. Connexin-26 mutations in deafness and skin disease. Expert Rev. Mol. Med. 11, e35 (2009).
Kelsell, D. P. et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387, 80–83 (1997).
Kelley, P. M. et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet. 62, 792–799 (1998).
Rabionet, R. et al. Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene. Hum. Genet. 106, 40–44 (2000).
Engel-Yeger, B. et al. The effects of a connexin 26 mutation–35delG–on oto-acoustic emissions and brainstem evoked potentials: homozygotes and carriers. Hear. Res. 163, 93–100 (2002).
Groh, D. et al. Hearing function in heterozygous carriers of a pathogenic GJB2 gene mutation. Physiol. Res. 62, 323–330 (2013).
Mutai, H. et al. Diverse spectrum of rare deafness genes underlies early-childhood hearing loss in Japanese patients: a cross-sectional, multi-center next-generation sequencing study. Orphanet J. Rare Dis. 8, 172 (2013).
Liberman, M. C. et al. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature 419, 300–304 (2002).
Jeong, G. et al. A Kelch domain-containing KLHDC7B and a long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer cell proliferation via the interferon signaling pathway. Sci. Rep. 8, 12922 (2018).
Martín-Pardillos, A. & Cajal, S. R. Y. Characterization of Kelch domain-containing protein 7B in breast tumours and breast cancer cell lines. Oncol. Lett. 18, 2853–2860 (2019).
Rudnicki, A. et al. Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. BMC Genomics 15, 484 (2014).
Liu, H. et al. Cell-specific transcriptome analysis shows that adult pillar and Deiters’ cells express genes encoding machinery for specializations of cochlear hair cells. Front. Mol. Neurosci. 11, 356 (2018).
GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Urbut, S. M., Wang, G., Carbonetto, P. & Stephens, M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 51, 187–195 (2019).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Carey, D. J. et al. The Geisinger MyCode community health initiative: an electronic health record–linked biobank for precision medicine research. Genet. Med. 18, 906–913 (2016).
Berglund, G., Elmstähl, S., Janzon, L. & Larsson, S. A. The Malmo diet and cancer study. design and feasibility. J. Intern. Med. 233, 45–51 (1993).
Dewey, F. E. et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354, aaf6814 (2016).
Van Hout, C. V. et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. https://doi.org/10.1038/s41588-021-00870-7 (2021).
Girotto, G. et al. Hearing function and thresholds: a genome-wide association study in European isolated populations identifies new loci and pathways. J. Med. Genet. 48, 369–374 (2011).
Nolan, L. S. et al. Estrogen-related receptor gamma and hearing function: evidence of a role in humans and mice. Neurobiol. Aging 34, e1–e9 (2013).
Wolber, L. E. et al. Salt-inducible kinase 3, SIK3, is a new gene associated with hearing. Hum. Mol. Genet. 23, 6407–6418 (2014).
Vuckovic, D. et al. Genome-wide association analysis on normal hearing function identifies PCDH20 and SLC28A3 as candidates for hearing function and loss. Hum. Mol. Genet. 24, 5655–5664 (2015).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Praveen, K. Colocalization locus zoom plots, figshare, https://doi.org/10.6084/m9.figshare.19233177 (2022).
Staples, J. et al. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 95, 553–564 (2014).
Delaneau, O., Zagury, J.-F., Robinson, M. R., Marchini, J. L. & Dermitzakis, E. T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 10, 5436 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
The authors would like to thank everyone who made this work possible. In particular: the UK Biobank team, their funders, the dedicated professionals from the member institutions who contributed to and supported this work, and most especially the UK Biobank participants, without whom this research would not be possible. The exome sequencing was funded by the UK Biobank Exome Sequencing Consortium (i.e., Bristol Myers Squibb, Regeneron, Biogen, Takeda, Abbvie, Alnylam, AstraZeneca, and Pfizer). This research has been conducted using the UK Biobank Resource under application number 2604. We also want to acknowledge the participants and investigators of the FinnGen study. We thank the MyCode Community Health Initiative participants for taking part in the DiscovEHR collaboration.
Author information
Authors and Affiliations
Consortia
Contributions
All authors contributed to critical review of the manuscript for important intellectual content, and final approval of submission of the manuscript for publication. Conceptualization: K.P., E.A.S., M.D., B.Z., A.B., G.C. Analysis: K.P., L.D., L.G., M.A.F., A.H.A., J.S., A.P., A.M., S.C., E.A.S., C.B., J.M. Single-cell sequencing data generation and analysis: S.M., Y.B., J.S. Mouse experiments: A.K. Project administration: E.C., M.J. Datasets: O.M. Supervision: M.A.F., G.A., E.A.S., S.B., B.Z., G.C., A.B. Writing – original draft: K.P., L.D., E.A.S., G.C. All authors contributed to securing funding, study design and oversight. All authors reviewed the final version of the manuscript. C.B., C.F., A.L., and J.D.O. performed and are responsible for sample genotyping. C.B, C.F., E.D.F., M.L., M.S.P., L.W., S.E.W., A.L., and J.D.O. performed and are responsible for exome sequencing. T.D.S., Z.G., A.L., and J.D.O. conceived and are responsible for laboratory automation. M.S.P., K.M., R.U., and J.D.O are responsible for sample tracking and the library information management system. X.B., A.H., O.K., A.M., S.O., R.P., T.P., A.R., W.S. and J.G.R. performed and are responsible for the compute logistics, analysis and infrastructure needed to produce exome and genotype data. G.E., M.O., M.N. and J.G.R. provided compute infrastructure development and operational support. S.B., S.K., and J.G.R. provide variant and gene annotations and their functional interpretation of variants. E.M., J.S., R.L., B.B., A.B., L.H., J.G.R. conceived and are responsible for creating, developing, and deploying analysis platforms and computational methods for analyzing genomic data. All authors contributed to the clinical informatics of the project. All authors contributed to the analysis of the project. All authors contributed to the management and coordination of all research activities, planning and execution. All authors contributed to the review process for the final version of the manuscript. All authors contributed towards the creation of the GHS-RGC DiscovEHR collaboration, helped frame research questions and contributed to the discussion and review of data and results, review and feedback on manuscript, and contributed to the management and coordination of discussions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: Regeneron authors receive salary from and own options and/or stock of the company. Decibel authors receive salary and may own options and/or stock of the company. The remaining authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Rick Friedman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Caitlin Karniski. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Praveen, K., Dobbyn, L., Gurski, L. et al. Population-scale analysis of common and rare genetic variation associated with hearing loss in adults. Commun Biol 5, 540 (2022). https://doi.org/10.1038/s42003-022-03408-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03408-7
This article is cited by
-
Sex differences in the polygenic architecture of hearing problems in adults
Genome Medicine (2023)
-
Rare-variant association analysis reveals known and new age-related hearing loss genes
European Journal of Human Genetics (2023)
-
Identification of hub genes associated with spermatogenesis by bioinformatics analysis
Scientific Reports (2023)
-
A prognostic and immunological analysis of 7B-containing Kelch structural domain (KLHDC7B) in pan-cancer: a potential target for immunotherapy and survival
Journal of Cancer Research and Clinical Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.