Abstract
DNA phosphorothioation (PT) is widely distributed in the human gut microbiome. In this work, PT-diet effect on nematodes was studied with PT-bioengineering bacteria. We found that the ROS level decreased by about 20–50% and the age-related lipofuscin accumulation was reduced by 15–25%. Moreover, the PT-feeding worms were more active at all life periods, and more resistant to acute stressors. Intriguingly, their lifespans were prolonged by ~21.7%. Comparative RNA-seq analysis indicated that many gene expressions were dramatically regulated by PT-diet, such as cysteine-rich protein (scl-11/12/13), sulfur-related enzyme (cpr-2), longevity gene (jnk-1) and stress response (sod-3/5, gps-5/6, gst-18/20, hsp-12.6). Both the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis suggested that neuroactivity pathways were upregulated, while phosphoryl transfer and DNA-repair pathways were down-regulated in good-appetite young worms. The findings pave the way for pro-longevity of multicellular organisms by PT-bacterial interference.
Similar content being viewed by others
Introduction
Phosphorothioate DNA (PT-DNA), in which a non-bridging oxygen in the phosphate link is replaced by a sulfur atom, is widely distributed among bacteria and archaea, according to the dndA-E functional gene cluster survey1. Similar to epigenetic methylation, the physiological DNA phosphorothioation is of specificity in Rp-chiral modification2,3 and GPSAAC/GPSTTC, GPSGCC, and CPSCA- consensus sequences (usually 12–14% frequency of these sequence motifs)4,5,6. At present, two biological functions of PT-DNA have been recognized by means of microbial studies: restriction-modification (R-M), to resist the invasive non-PT-modified genetic material of bacteriophages7,8,9; and on-site DNA protection, to counteract ROS assault10,11,12. The R-M system is constrained to the bacterial compartment in the case of intracellular gene-cluster mechanics (either dndF-H or pbeA-D)8,13,14. However, the anti-ROS “one-atom S/O chemistry” property can be feasibly inherited when other organisms ingest PT-containing bacteria11,12,15.
Typically, the S/O anti-ROS mechanism suggests that ROS molecules could be transformed to protective thiyl species whenever phosphorothioate is oxidized to normal phosphate12. Besides the straightforward redox function, a combinative analysis of epigenome, transcriptome, and metabolome has indicated that PT modification also contributes to the cellular redox state and stress resistance in bacteria1. Intriguingly, Wu et al. discovered physiological phosphorothioate RNA in both prokaryotes and eukaryotes, e.g., HeLa cells, with highly sensitive liquid chromatography coupled with tandem mass spectrometry16. Recently, a human microbiome study showed that over 2000 PT-gene-containing bacterial strains, including the genera Pseudomonas, Clostridioides, and Escherichia, were distributed in the gastrointestinal tract and six PT-dinucleotides (CPSG, CPST, APSG, TPSG, GPSC, and APST) were detected in human fecal DNA17. These discoveries demonstrate the existence of DNA/RNA phosphorothioation in both prokaryotes and eukaryotes, and it is worthwhile to understand the possible long-term diet effects of PT-containing bacteria.
Currently, many anti-ROS phenotypes and molecular mechanisms have been investigated for PT-containing bacteria. Wang et al. have reported that catalase and organic hydroperoxide resistance gene expressions were not up-regulated in the PT-containing wild type Streptomyces lividans18, though the PT-containing bacteria exhibited 2 to 10-fold higher survival under peroxide oxidative stress and its ROS scavenging was much faster than the Dnd- mutant. Furthermore, PT modification plays a dual role in protecting and damaging genome—the PT site sacrifices itself to generate a sulfur-derived ROS scavenger, but simultaneously increases the chance of DNA fragmentation at the PT site during the antioxidative process12,19. The fragmentation-after-acting characteristic promotes PT-containing DNA fragments release from dead bacteria to environment under oxidative stress, and increases its intake by surrounding bacteria or as nutrients ingested by host organisms. Beyond anti-oxidative stress, PT-containing bacteria also exhibited resistance to heavy metals, and to ultraviolet and X-ray radiation11,12,18,20. Particularly, previous studies have shown that phosphorothioate plays a mild protective role, without pro-oxidative side-effects11. It is worth knowing whether the host can benefit from the long-term diet intake of phosphorothioate as well.
In this study, we fed the wild-type C. elegans strain (Bristol N2) either PT-bioengineered E. coli OP50 or a control. Then the ROS level and lipofuscin accumulation, body and brood sizes, locomotion, and lifespan were compared between the two groups of nematodes, and RNA-seq analysis was carried out at the most discriminating time points (four-day and twelve-day old, D4 and D12) to discern any change in gene-expression. It was observed that the ROS level substantially decreased in C. elegans fed PT-containing bacteria, and the worms became healthier in terms activity and longevity. Furthermore, the PT-fed worms exhibited tolerance to environmental stresses caused by paraquat, heat, and Cr6+, but not for ultraviolet radiation. Our results suggest that C. elegans ingest phosphorothioate from PT-bacterial food. Unlike the bacterial transcriptome18, the C. elegans transcriptome exhibited statistically meaningful regulation of the expression of many genes, including but not limited, to neuroactivity, DNA repair, phosphoryl transfer, defense-response, and sulfur-related metabolism. It is likely that the bacteria-sourced phosphorothioates take part in multiple pathway-network regulations and thereby influence the observed phenotypes, such as locomotion and aging.
Results
PT-containing bacteria food reduces the ROS level in C. elegans
The most remarkable and straightforward long-term diet effect was on the ROS level in C. elegans. Figure 1 compares the ROS levels and lipofuscin accumulation of C. elegans fed with E. coli OP50 harboring a pBluescript-based plasmid (pJTU1238) (S + , Dnd phenotype positive), which has the dnd gene cluster from Salmonella enterica, and normal E. coli OP50 (S-, Dnd phenotype deficient, negative control). Plasmid DNA from the two E. coli OP50 strains were tested by using iodine-induced PT-DNA cleavage to confirm PT-modification in the bacteria12. To examine the long-term effect of exogenous phosphorothioate on C. elegans, the wide type Bristol N2 nematodes were fed E. coli OP50 S + (Dnd + ) and normal OP50 (S-, Dnd-).
a ROS level measured by the fluorescent probe H2DCF-DA (excitation, 488 nm; emission 550-600 nm, nS+ > 35, nS- > 36); b ROS level measured by the fluorescent probe DCM (excitation, 478 nm; emission 550-600 nm, nS+ > 28, nS- > 27); and c) Autofluorescence of lipofuscin (excitation, 478 nm; emission 630–700 nm, nS+ > 24, nS- > 26). C. elegans samples were collected on Day 1, 4, 8, and 12, after L4 larva-to-adult molts at 22 oC, the images with fluorescence close to average reads were selected as examples in Fig. 1. Central lines represent the mean values, and error bars note the standard deviation (rfu, relative fluorescence units. Statistical significance was calculated with two-tailed t-test and p values are indicated. see the details in Supplementary Figs. 1–3).
As shown in Fig. 1a and Supplementary Table 1, the H2DCFDA-related ROS level decreased by 18.8, 19.1, and 32.7% at day 1, 4, and 8, respectively, while it slightly decreasing by 0.9% at day 12. Likewise, the DCM-related level respectively decreased by 19.9, 54.0, and 21.5% at day 1, 4, and 8, but no significant change at day 12. The p-values for the lower ROS levels in the PT-fed nematodes were less than 0.001 in the D4 and D8 samples by the DCM measurement, and less than 0.01 by the DCF measurement. No statistically significant ROS drop was observed for the D1 and D12 samples.
The 2′, 7′-dichlorodihydrofluorescein diacetate (H2DCFDA) fluorescent probe passively diffused into cells and reacts with most ROS species, including hydrogen peroxide, the hydroxyl radical, and peroxynitrite after cleavage by in vivo esterase21,22. Upon oxidation by an ROS species, the nonfluorescent H2DCFDA was converted to 2′, 7′-dichlorofluorescein (DCF), and thereby, the DCF fluorescent intensity could be a linear response to the ROS molecules in C. elegans23,24,25. Similarly, the nonfluorescent 2-(2-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)styryl)-4H-chromen-4-ylidene)malononitrile (i.e., DCM boronate) was converted to fluorescent dicyanomethylene-4H-pyran fluorophore; but the DCM fluorogenic probe was more sensitive to hydrogen peroxide and peroxynitrite26,27. Interestingly, ROS quantification appeared more dramatic when measured by DCM for C. elegans fed with PT-containing OP50, even though no NOS gene (to biosynthesize nitric oxide, a common peroxynitrite precursor combining with superoxide) existed in N2 C. elegans or E. coli OP5028.
ROS suppression by PT-containing bacterial diet was in a good agreement with previous studies of phosphorothioate antioxidant properties10,11,12. Typically, the S/O anti-oxidant mechanism is also ROS species-related—the PT-genome is highly sensitive to some ROS species like hypochlorous acid19, whereas phosphorothioation is more protectable in the presence of peroxynitrite and hydrogen peroxide11,12. One rationale is that competitive P-O and P-S cleavages occur upon ROS attack. If the double-stranded P-O cleavage occurs on the PT site, then the bacterial genome would be fragmented and apoptosis. On the other hand, if only the P-S bond is broken by a ROS species, then the resulting thiyl derivatives would scavenge the surrounding ROS species. For an extreme example, iodine (I2) can effectively crack the bridging P-O bonds at the PT-modified site of PT-DNA and is widely used to detect the PT-DNA phenotype5. Hydrogen peroxide and peroxynitrite prefer the PS/PO conversion via P-S cleavage at the PT-modified site. Thus, the low ROS level in the PT-fed nematodes indicates that phosphorothioate may act as an anti-ROS species after digestive intake.
Figure 1c shows the age-accumulation of fluorescent material in C. elegans, which was usually considered as autofluorescent lipofuscin from the aging process. In wild-type animals, age pigments increase into adulthood, accumulating slowly during the reproductive phase and more rapidly during the post-reproductive period29,30,31. Thus, lipofuscin accumulation indicates a physiologically aged state, rather than simply marking chronological time. The lipofuscin accumulation of the nematodes fed with PT-containing bacteria was much slower than those with normal OP50 diet. Therefore, a delayed rate of aging coincides with the consumption of PT-containing food, according to the lipofuscin biomarker measured by autofluorescence.
Phenotype changes potentially caused by PT-containing food
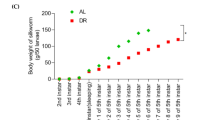
The locomotion, lifespan, body, and brood sizes of C. elegans (Bristol N2 strain) post L4-to-adult molt are illustrated in Fig. 2, with comparison of PT-containing bacterial food and normal OP50 food.
a The percentage of nematodes with class a motility ((S+)-fed C. elegans were more active; p-value < 0.01, nS+ = 78, nS- = 75). b Percentage alive in the two groups of worms, observed from the nominal age of 3 days ((S+)-fed C. elegans had longer lifespans; p-value < 0.01, nS+ = 132, nS- = 124). c Body sizes of the two groups of nematodes (no significant change; nS+ > 14, nS- > 20). d Brood sizes of the two groups of nematodes (no significant change; nS+ = 8, nS- = 8). p-values were calculated with log-rank test.
The motility of worms was classified into four ranks: (a) robust, coordinated sinusoidal locomotion; (b) uncoordinated and/or sluggish movement; (c) no forward or backward movement, but head movements or shuddering in response to prodding; and (d) dead animals32,33. Figure 2a shows the percentage of class a—those that exhibited spontaneous movement or vigorous locomotion in response to prodding (also see the percentages of classes b–d in Supplementary Fig. 4). The worms fed with PT-containing bacteria were much more active, particularly during day 10 to 16 (class a difference: 19.9 ± 3.0%). All the motility scores of animals fed with PT-containing food were equal to or higher than control at every age tested.
The percentage of nematodes remaining alive in the two groups were recorded under normal culture conditions (ntotal > 250). The lifespans of the animals fed with Dnd + E. coli OP50 (S+) were significantly longer than those fed with normal E. coli OP50 (Fig. 2b and Supplementary Table 2). The mean lifespan of the two groups of worms were 20.2 ± 1.4 and 16.6 ± 0.8 days respectively (those fed PT-containing bacteria had a 21.7% longer lifetime). To ensure the rigor of the experiment, long-lived (daf-2, n = 117) and short-lived (daf-16, n = 114) C. elegans were cultured under the same conditions with normal E. coli OP50 diet. The resulting lifespans were 30 and 13 days on average, respectively (p-value < 0.001, see Supplementary Table 2 and Supplementary Fig. 5). Thus, the exogenic effect of food (~3.6 day) on lifespan was much less than the genetic effects (e.g., daf-2 mutants live longer by 13.7 days, and daf-16 shorter by 3.4 days under the same experimental condition). Heat-killed bacteria have the similar diet effect on the lifespans of nematodes, confirming the PT-diet effects on their longevity and also suggesting PT-bacteria function as food rather than live gut microbe. (See Supplementary Tables 3–4 and Supplementary Figs. 6 and 7.) Even though both the alive and heat-killed bacteria prolong nematodes’ lifespans more or less, the degree of lifespan change is different: for the long-lived daf-2 mutant, the PT-diet enhancement is 5.0 and 4.3% with the alive and heat-killed bacteria, respectively (p-value, 0.01–0.02); for the short-lived daf-16 mutant, the PT-diet enhancement is 13.6 and 13.0%, respectively. (p-value, 0.003–0.03).
As shown in Fig. 2c–d, the body and brood sizes of the two types of nematodes were not significantly different. The biological-engineering diet intervention had little effect on body development.
Taken together, the above phenotypic findings indicate that PT-containing bacterial food was coincident with increased motility and longevity of C. elegans, but they aren’t having more offspring and their body sizes are the same.
Stress responses for exogenous stimulus
A variety of studies have demonstrated that various kinds of stressors exert deleterious effects by increasing ROS production20,34,35,36,37. It is worth testing whether the diet effect is also protective in cases of acute oxidative injury. As shown in Fig. 3a–d, paraquat, K2Cr2O7, heat shock, and ultraviolet radiation were used to evaluate responsiveness to acute oxidative stress in C. elegans. The survival rates were recorded hours after the stresses imposed upon the two groups of nematodes.
Survival percentages after treatment with (a) paraquat ((30 mM nS− = 138, nS+ = 146, p-value < 0.05), (b) Cr6+ ((10 mM nS− = 111, nS+ = 109, p-value < 0.05), (c) heat shock (35 °C for 6 h, nS- = 127, nS+ = 136, p-value < 0.01), and (d) ultraviolet irradiation (nS- = 41, nS+ = 38, p-value < 0.001). Animals were exposed to the stresses on day 3 after L4-to-adult molts, and p-values were calculated with the log-rank test.
The mean lifespans of C. elegans S+and C. elegans S− were 93.3 ± 31.8 and 83.2 ± 20.0 h after paraquat poisoning, and 71.5 ± 31.6 and 60.9 ± 21.8 h after Cr6+ poisoning, respectively (see Supplementary Table 5). The PT-containing diet may have helped in the cases of paraquat and Cr6+, (survival increased by 12.1 and 17.4% on average, respectively. p-value < 0.05), and more so against heat shock (17.4 %, p-value < 0.01). However, there may be adverse side-effects associated with exposure to ultraviolet radiation while on the long-term S+diet (survivals decrease by 15.6%, p-value < 0.001).
Survival percentages associated with long-term consumption of PT-containing bacterial food were likely related to the antioxidant effects of phosphorothioate compounds (see Supplementary Table 6 and Supplementary Fig. 8). Under the same chemical stresses caused by paraquat, if in the presence of phosphorothioate diethyl ester (DEPT), the survival percentages increased significantly by 23.7 as well—but DEPT is noneffective for Cr6+-induced stress. Interestingly, PT-containing plasmids were photo-sensitive and underwent photolysis whereas Dnd + bacteria appeared to have stronger resistance to ultraviolet radiation compared to Dnd- bacteria (see Supplementary Fig. 9)—the self-sacrifice of PT-DNA protected the neighboring organisms from the UV damage. This duality implied that PT-containing diet can help C. elegans resist certain ROS species, but perhaps not all of them.
Thus, it is quite reasonable to assume that the low ROS level in PT-fed nematodes stemmed from the long-term intake of phosphorothioate, which also helps to prolong the lifespan and promote their motility. However, the intake of phosphorothioate may lead to more complicated effects in C. elegans, at the level of the transcriptome and metabolome.
Comparative transcriptome analysis of C. elegans fed PT-containing food
Since phenotypic investigation revealed the most essential changes in young and middle-aged nematodes fed with PT-containing OP50, RNA sequencing was performed on the 4th and 12th day after adulthood, as representative of the youth and early aging stage, respectively38.
As shown in Fig. 4a, each group of samples gave high-quality FPKM (fragments per kilobase of transcript per million mapped reads) in RNA-seq. The PT-feeding groups presented relatively higher FPKM values in a range of 22–1 to 25–1, in particular for the D4 samples (see Supplementary Fig. 10). In Fig. 4b and Supplementary Table 7, compared to the non-PT-fed nematodes, more genes were transcribed in those that were PT-fed, in both the D4 and D12 samples (see the detailed D4–D12 comparisons in Supplementary Figs. 11–16 and Supplementary Tables 8–14). The number of differentially expressed genes (DEGs) in the D4 samples were more than those in the D12 samples, especially for down-regulated genes. On average, 2271 and 1931 genes were upregulated by nematodes that consumed PT-containing bacterial food in the D4 and D12 samples (|Log2FoldChange| > 1 and adjusted p value padj < 0.01), respectively. On the other hand, 2891 and 380 genes were down-regulated at the same time. Only 234 of the upregulated genes and 115 of the downregulated genes overlapped between the D4 and D12 samples, indicating the PT-containing food had different effects depending on the age of C. elegans39,40. It is likely that long-term PT-containing food affects young C. elegans more significantly than the middle-aged worms.
In particular, a few outlier genes with extremely small padj-values and extremely large fold changes are listed in Table 1.
Intriguingly, the biological function of top-ranked up- and down-regulated genes pointed to many cysteine-related (i.e., sulfur-related) enzymes and lifespan-related daf-2/16 genes; their regulator genes such as cep-1, pal-1, and vrk-1 are systemically down-regulated while gbb-1 is up-regulated. (See Supplementary Table 15.) It was very rare to observe genes that were up- or down-regulated by more than 50 times in gene expression, such as scl-11/12/13 (cysteine rich protein) and Y41C4A.32 (daf-2 related) transcripts. In a recent study of mitochondrial longevity regulated by modulating ER genes, transcript Y41C4A.32 was affected by isp-1 and nfyb-141 and thereby involved in daf-2/insulin-IGF signaling of long-lived mutants. In this work, Y41C4A.32 was down-regulated by ~50 fold in the D4 samples, but slightly up-regulated by 1.3-fold in the D12 samples, reflecting a significant fluctuation during aging. To date, we do not know whether these changes in the transcriptome were directly connected to phosphorothioate as in antisense drugs or not, for lack of detailed information about phosphorothioate nucleotides in the organism.
Gene expression related to the stress responses and aging
Inspired by the RNA-seq overview analysis42,43,44,45,46, we focused on the D4 samples to mine clues about anti-ROS and anti-aging functions.
As shown in Fig. 5a–h and Supplementary Table 16, sod-3, sod-5, gpx-5, and gpx-6 were increased by 10.3, 16.7, 2.3, and 2.3-fold in the C. elegans S+/D4 samples compared to the C. elegans S-/D4 (control). In addition, the expression of heat shock protein-12.6 (hsp-12.6) increased by 9.7 and 5.3-fold in the D4 and D12 samples, respectively. It was known that sod, gpx, and hsp genes were related to antioxidant functions. This suggested that bacterial phosphorothioate not only scavenged ROS in C. elegans directly, but also activated these anti-ROS genes in the worms. It should be noted that the anti-ROS gene overexpression is unlikely caused by the reduced ROS level, probably due to the complicated global transcriptomic regulations47,48,49. Interestingly, many expressions of glutathione S-transferases (GST) genes were induced in both the D4 and D12 samples (see Fig. 5f), indicating that the long-term intake of PT-containing food may change sulfur metabolism in the nematodes and induce the expression of GST, which may lead to life extension by increasing antioxidant activity.
a, b Venn diagrams of up- and down-regulated genes of the Caenorhabditis elegans, using S-/D4 as a reference for the other three groups of samples. c Fold-change heatmap of overall gene clustering. d–h Fold-change heatmaps of the genes for ROS-scavenging, heat-shock protein, glutathione S-transferase, age resistance and promotion for C. elegans, with reference to the C. elegans S-/D4 samples.
Figure 5g–h shows a few aging-related genes with different expressions in the PT-fed and control groups. The stress-response gene jnk-1 increased by 2.2-fold and the aging-promoting gene glp-1 decreased by 2.9-fold, which might be related to the change in lifespan. Indeed, the aging-related gene analysis indicates the insulin/IGF-1 (IIS) and dietary restriction pathways might involve in the PT-induced longevity effect, because PT-regulated expressions of mtl-1, col-10, col-13, daf-12, jnk-1, pha-4, ttx-1, ser-3, daf-18, mes-4, ins-7, sams-1, and glp-1 are anti-aging (see Supplementary Table 17).
Quantitative real-time PCR (qRT-PCR) validated PT-diet regulation on gene expressions discovered by the RNA-seq results. The regulatory patterns of scl-11, scl-12, scl-13, ZK218.3, cpr-2, Y41C4A.32, T22B7.7, C44B7.5, sod-3, sod-5, gpx-3, gpx-5, gpx-6, cat-1, cat-2, hsp-12.6, gst-10, gst-15, gst-18, gst-20, gst-28, jnk-1, glp-1 measured by the qRT-PCR were in agreement with the transcriptomic changes measured by the RNA-seq method (correlation coefficient, R2 = 0.9476) (see Supplementary Figs. 17 and 18 and Table 18).
Thus, PT-containing bacterial food may act as antioxidant directly to oxidative stress and substantially change many gene expressions by the involvement in the nematodes’ transcriptomic network. Interestingly, certain antioxidant enzymes like SODs and GSTs were overexpressed significantly, different from the reduced expression in PT( + ) bacteria under oxidative stress18.
Global gene ontology (GO) enrichment of PT-feeding C. elegans
Figure 6 illustrates the GO enrichment of the differences between the transcriptomes of C. elegans S+/D4 and C. elegans S-/D4 samples. Overall, 101, 383, and 123 categories were enriched in ‘cellular component’, ‘biological process’, and ‘molecular function’, while only 20, 43, and 26 categories were enriched in the corresponding D12 samples (Supplementary Data 1–4). This indicates that the long-term PT-containing food had a greater impact on young nematodes, which may have been caused by the nematodes eating more food at a young age. For clarity, the top-rank differential GO terms are listed in Table 2.
The GO Directed acyclic graph (DAG) shows the enriched GO terms in the downregulated cellular component, biological process, and molecular function categories. The boxes and circles represent GO terms and are labeled with the GO ID on the first line, the term definition and information on the second line and the p-value on the third line. The fourth line is labeled with the number of regulated genes associated with the listed GO ID and the total number of genes. Annotation moves from more general to more specific with progression from the parent nodes. The top-ranked terms with the lowest p-values are marked with boxes and yellow/red colors. The more red-saturated a node is, the greater its significance.
As shown in Table 2, a few genes with phosphoryl transfer-related GO terms (0018107, 0006470, 0004721, and 004725) had statistically significant regulation. It appears likely that phosphorothioate is involved in phosphoryl-transfer processes in the nucleus, e.g., phosphorylation and dephosphorylation of enzymes and kinases. Intriguingly, for some reason, most of the regulations induced by PT-containing food were to suppress the expression of phosphoryl-transfer genes. The PT-containing diet probably alleviated DNA replication stress by its anti-ROS activity, which might result in less cellular damage50,51,52. So, the cell cycle can run at a relatively low rate without changing worms’ development and growth. It is not clear how (or even, if) phosphorothioate suppresses gene expressions at the DNA–RNA transcription level.
Since the top-ranking GO terms were dominated by down-regulated genes that had much lower padj values than up-regulated genes, we specifically checked the top-three GO terms corresponding to gene upregulation. Those top three GO terms in each of the three GO aspects were collagen trimer, ciliary part, and cell projection part in the ‘cell component’ class; neuropeptide signaling pathway, phototransduction, and detection of light stimulus in the ‘biological process’ class and structural constituent of cuticle, oxygen binding, and tetrapyrrole binding in the ‘molecular function’ class. Owing to the poor padj-values, the top-ranking GO terms may change if only down-regulated genes were considered. Among them, the upregulation on neural signaling pathways may connect the high motility of the worms fed with PT-containing food, the oxygen and tetrapyrrole binding may relate to P450-like genes, and the phototransduction and detection of light stimulus (probably on cuticle and ciliary parts) may be related to ultraviolet sensitivity.
In the D12 samples, defense response, innate immune response, immune response, immune system process, and defense response to other organisms were the top five categories attributed to ‘biological process’ (Supplementary Data 4). Structural molecule activity, structural constituent of cuticle, carbohydrate binding, peptidase activity, acting on L-amino acid peptide activity, and protein dimerization activity were the top five categories in the ‘molecular function’ aspect. Immune response and protein dimerization were reduced, probably the PT-fed nematodes were aging more slowly and less aging-related immune response. The overall differences in the D12 samples were much smaller than those in the D4 samples, and no specific loci of the ‘cell component’ aspect was captured in the GO analysis of the D12 samples. The most significant change is phosphoryl-transfer terms (GO: 0031143/0018107/0018210/0006470 /0004721/0004672) that were downregulated at day 4 and upregulated at day 12, which may be related to the increasing motility at day 12 (see Supplementary Table 19).
KEGG pathway enrichment of PT-feeding C. elegans
The Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses for DEGs of S+/D4 compared S-/D4 are shown in Fig. 7. Among the top-three regulated pathways, both the DNA repair (cel03030) and mismatch repair (cel03430) pathways were down-regulated (padj = 1.4 × 10−8 and 1.3 × 10−6), but neuroactive ligand-receptor interaction (cel04080) was up-regulated (padj = 9.6 × 10−7). They may be correlated with the PT-DNA self-repair mechanism and the high motility observed in the phenotypes.
As shown in Fig. 7b–c, the most dramatic changes in the top-ranked regulation pathways were in a range of 2 to 3-fold, except for lgc-36 (~5.2 fold), tkr-2 (~4.7 fold), T24C4.5 (~3.9 fold), and mcm-7 (~3.1 fold). lgc-36 is a protein-coding gene of the gamma-aminobutyric acid receptor (GARP, α-subunit), with corresponding transcript F07B10.5.1 (17/34 GAAC/GTTC, 6476 bp). tkr-2 is a tachykinin G protein-coupled receptor gene, with corresponding transcript C49A9.7.1 (21/31 GAAC/GTTC, 7823 bp). T24C4.5 coded DNA primase (54/56 GAAC/GTTC, 13349 bp). mcm-7 is a DNA replication licensing factor gene, with corresponding transcript F32D1.10 (20/22 GAAC/GTTC, 6609 bp). Six of seven mcm helicase gene expressions decreased by 2.6-fold on average, which may be connected to the phosphorothioate destabilization of the B-type DNA helix3,6. There are at least two possible reasons for the lowered DNA replication and mismatch repair genes expressions, one could be less oxidative DNA damage due to a relatively low ROS level, and another could be the higher flexibility of double and triple-stranded helices in helicases upon phosphorothioate involvement. Global regulation of transcripts with similar multiplicative changes suggests that phosphorothioate systemically targets gene networks, rather than specific genes.
The other up-regulated pathways included neuroactive ligand-receptor interaction, calcium signaling pathway, purine metabolism, drug metabolism - cytochrome P450, metabolism of xenobiotics by cytochrome P450, and glutathione metabolism in the D4 samples, while only biosynthesis of amino acids, cysteine, and methionine metabolism pathways were upregulated in the D12 samples (Supplementary Data 5–6). Basically, the KEGG enrichment pattern was consistent with the GO enrichment pattern concerning upregulation of neuroactivity.
Interestingly, glutathione metabolism was upregulated in the D4 samples and many glutathione S-transferase (GST) genes were induced, consistent with the differential gene expression in Fig. 4. This suggests that PT-containing food would change sulfur-related metabolism in organisms.
The other downregulated pathways were the fanconi anemia pathway, homologous recombination, nucleotide excision repair, base excision repair, Wnt signaling pathway, ubiquitin mediated proteolysis, biosynthesis of amino acids, and the TGF-beta signaling pathway in the D4 samples, while no specific pathways were meaningfully downregulated in the D12 samples (Supplementary Data 7).
Discussion
The overall PT-containing bacterial food effects on C. elegans were integrated in Fig. 8. Basically, Dnd + bacteria brought their unique phosphorothioate materials into worms’ intestinal tract. Ultimately, bacteria were digested into small nutrient molecules, including phosphorothioate and their nucleotide oligomers. The long-term diet intake of phosphorothioate could help young nematodes to maintain a relatively low-level of ROS stress in their body, enabling higher locomotion and extending lifespan. The superior phenotypes could also contribute to the complicated regulation of gene expression by the phosphorothioate nutrients from the long-term Dnd + bacterial food, besides S/O anti-ROS activity.
The nematodes (pink-colored) uptakes dnd-gene bioengineering E. coli OP50 (brown-colored), in which plasmid and genome DNA undergo phosphorothioation by Dnd function proteins. The bacterial PT-DNA is thus ingested by the nematodes, converting ROS to reductive species via sulfur-oxygen one-atom chemistry. Simultaneously, the bacterial PT-DNA leads to globally interference on gene expressions of scl-11, 12, 13… The system-biological regulation is characterized by down-regulation of phosphoryl transfer GO terms and DNA repair/replication in KEGG enrichment, but up-regulation of neuroactivity. The overall effects enhance stress-resistance, enable locomotion, and extend lifespan.
The physiological roles of reactive oxygen species (ROS) can be quite complicated, and also divergent. ROS molecules are generated in aerobic metabolism, regulating many important cellular signaling transmissions; however, their over-generation causes oxidative damages, which is detrimental to cell function53,54,55. One side-effect of ROS accumulation during aerobic respiration is accelerated aging56,57,58. Traditionally, cellular antioxidants such as high-concentrations of vitamin C and free thiols (e.g., cysteine and glutathione) easily increase oxidative stress, namely prooxidativity59,60,61,62. The mild antioxidant phosphorothioate successfully avoids the prooxidative problem and is considered to be a high-quality antioxidant that only regulates the cellular ROS level to be within a healthy range11,12. In this study, we also observed a significant ROS-level decline measured by the H2O2 and peroxynitrite- specific probe, DCM. Even though C. elegans lacks a nitric oxide synthase gene, such that its ability to form peroxynitrite is questionable, bacterial nitric oxide is a nematode hormone28 and might be involved in peroxynitrite formation in the presence of superoxide.
Beyond the straightforward S/O anti-ROS mechanism, the regulation of gene expression of antioxidant enzymes such as superoxide dismutase, catalase, glutathione reductase, and glutathione peroxidase were stunning63,64,65,66. Previous bacterial transcriptome studies presented a paradox concerning whether the redox state of Dnd + bacteria was affected by PT-modification or not1,18. The bacterial investigations in literature have in part been inconclusive due to the dramatic difference of genetic backgrounds from Streptomyces lividans to E. coli B7A, conflating gene-expression caused by PT-modification with that of the genetic baseline. In this work, the genetic background of the C. elegans N2 strain was identical for the two established groups of nematodes. Accordingly, gene-expression regulation became very stable and redox-related genes, like SODs and GSTs, were successfully identified.
Another interesting discovery was the potential enhancement of locomotion and neuroactivity by consumption of PT-containing food. To the best of our knowledge, support for the upregulation of neural signaling pathways was found for the first time, and affected worms retained the motility significantly longer. The molecular mechanism for phosphorothioate-neuroactivity has not been determined, but is a very promising avenue of future investigation. Particularly, the body size (an equivalent index of body weight) remained unchanged, making phosphorothioate a potential treatment for neurodegenerative diseases such as Alzheimer’s disease.
Indeed, phosphorothioate oligonucleotides have been long used in antisense drugs for clinical research since the 1970s67,68,69,70,71, for that phosphorothioate can slow down phosphoryl hydrolysis and thereby retain pharmacokinetics. Antisense drugs target many diseases, including neurodegenerative disorders, cancers, infections, and other diseases68,70,71. It is worth revisiting phosphorothioate’s cryptic function as anti-oxidant and neuro-activator.
Accordingly, the microbiomics roles of Dnd+ bacteria in human gut appear auspicious. Phosphorothioation was promoted under many stresses20, and thereby Dnd+ bacteria may increase the PT-modification frequency on their genomic materials. On the other hand, the chance for bridging P-O cleavage arises during the PS/PO conversion, which leads to genomic instability and bacterial death. Phosphorothioate release could help lower the surrounding ROS level for neighboring bacteria and the host organism. The different ROS level detected by the two fluorescent probes implies that ROS species were not evenly declining upon the PT-containing food, and in particular, hydrogen peroxide may decrease more significantly than other ROS species.
Finally, increased longevity could be one of the best discoveries from PT-fed C. elegans. Lifespan extension was confirmed by age-related lipofuscin attenuation. PT-diet upregulated 64.3% of Class I (4178 detected in this work) and downregulated 61.1% of Class II (3996 detected) in the over 18,000 daf-16 target genes (see Supplementary Fig. 19, also see the extending comparisons in Supplementary Figs. 20–21), indicating that PT-diet pro-longevity might share similar signaling pathways with daf-16 such as insulin/IGF-1 and dietary restriction pathways—the global regulation on gene expressions could be implemented by bacterial phosphorothioate-involving core sequence elements like DBE (daf-16 binding element) and DAE (daf-16 associate element)72. Multiple factors may influence prolonged life in C. elegans, including a lowered ROS level, suppressing phosphoryl transfer, alleviating DNA repair burden, and regulating redox enzyme and protein levels. The pro-longevity role of PT-diet is milder than that of daf-2/16 mutations, but is more practical for pharmaceutical application.
Methods
Strains and culture conditions
Escherichia coli OP50, which is used as the standard food for C. elegans cultivation, was grown on nematode growth medium (NGM) at 37 °C. Plasmids with dndC-E gene cluster were transformed to E. coli OP50 to construct a PT-DNA containing strain named E. coli OP50 (S+). Wild-type N2, short-lived mutant (daf-16)73 and long-lived mutant (daf-2)74 worm strains were received by professor Wang Da-Yong from Southeast University. Worms were maintained on NGM plates (1.7% agar, 2.5 mg/ml peptone, 25 mM NaCl, 50 mM KH2PO4, pH 6.0, 5 μg/ml cholesterol, 1 mM CaCl2, and 1 mM MgSO4) kept at 25 °C and spotted with E. coli OP50 or E. coli OP50 (S+) as the food source.
Generalized assay preparation
Age-synchronization was accomplished by allowing ten young adult worms lay eggs on a fresh NGM plate with OP50 (S−) or OP50 (S+) for 4 h at 25°C. Adult worms were then removed and the plates were incubated at room temperature (25 oC) for 3 days75.
Measurement of Reactive Oxygen Species (ROS) in Caenorhabditis elegans
Worms were grown on NGM plates containing OP50 or OP50 (S+) bacteria, and then subjected to ROS assays. ROS formation in C. elegans was measured using the fluorescent probes 2,7-dichlorodihydrofluorescein diacetate (H2DCF-DA)23,24,25 and 2-(2-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)styryl)-4H-chromen-4-ylidene)malononitrile (DCM)26,27. The animals cultured at the appropriate time (1, 4, 8, or 12 days of adulthood) were collected into 1 mL phosphate-buffered saline (PBS) in Eppendorf tubes and washed 3 times with fresh PBS, then the nematodes were incubated with 50 μM H2DCF-DA and DCM for 60 min at room temperature separately, while the control was incubated with the same volume of DMSO. After that, animals were all washed 3 three times with fresh PBS and placed onto a slid with 3% agar plate containing 2 mM levamisole59. DCF Fluorescence intensity was measured at excitation and emission wavelengths of 488 and 510–560 nm, respectively, by means of laser scanning confocal microscopy (Leica & TCS SP8)23,24,25. The fluorescence intensity of DCM was measured by laser scanning confocal microscope (Leica & TCS SP8) at excitation and emission wavelengths of 488 and 630–750 nm, respectively26,27. The assay was performed in three independent trials, and at least 25 animals were used in each assay.
Lipofuscin analysis
Worms were grown on NGM plates containing OP50 or OP50 (S+) bacteria, and then subjected to lipofuscin analysis. The autofluorescence of intestinal lipofuscin was measured as an index of senescence at day 1, 4, 8, and 12 of adulthood. Randomly selected worms from the plates with OP50 and OP50 (S+) were washed three times with M9 buffer. Worms were then placed onto a 3% agar plate containing 2 mM levamisole. Lipofuscin autofluorescence was detected by laser scanning confocal microscope at excitation and emission wavelengths of 478 and 550–600 nm, respectively (Leica & TCS SP8)29. The assay was performed in three independent trials and at least 20 animals were used in each assay.
Measurement of locomotion
Synchronized three-day-old worms were placed on mNGM plates covered with lawns of OP50 and OP50 (S+). The motility of the worms was then examined using the scoring method described in previous studies33. Briefly, worms were placed on mNGM plates supplemented with 0.1 mM of FUdR (2′-deoxy-5-fluorouridine) and covered with lawns of OP50 and OP50 (S+) classified as class “a” when they exhibited spontaneous movement or vigorous locomotion in response to prodding; class “b” worms were those that did not move unless prodded or appeared to have uncoordinated movement; and class “c” worms were those that moved only their head and/or tail in response to prodding. Dead worms were classified as class “d”. Three independent replicates were tested, and the merged data were statistically analyzed and presented.
Lifespan assay
Age-synchronization was accomplished as described above34. >90 worms were randomly selected from both groups and transferred to three fresh NGM plates with live OP50 or OP50 (S+), correspondingly, and supplemented with 0.1 mM of FUdR to avoid progeny. The animals were transferred daily to fresh plates for the first four days, and thereafter, they were transferred every other day until all nematodes were dead74. Animals were scored daily by gentle prodding with a platinum wire76. Those animals that failed to respond to a gentle touch with a sterilized platinum wire were scored as dead75,76. Animals that crawled off the plate or died from internal hatching were censored. Worm survival was calculated by the Kaplan–Meier method, and differences in survival were analyzed using the log-rank test. All experiments were performed at 25 °C, except for the heat shock treatment.
Measurement of body size
Age-synchronized nematodes were prepared as describe above, then grown on NGM plates covered with lawns of OP50 or OP50 (S+) until the late larval stage, L4 (3 days). >90 adult worms were placed on 3 fresh plates with OP50 or OP50 (S+), and supplemented with 0.1 mM of FUdR to avoid progeny. The plates were incubated at 25 °C, and the body size of the live worms was measured every day until the worms reached the age of 10 days. Images of nematodes were taken with a stereoscopic microscope (Shanghai Bimu & XTL-3400E) equipped with a camera and analyzed by ImageJ software. The areas of a worm’s projection were used as an index of its body size.
Measurement of brood size
Age-synchronized nematodes were prepared as if for a lifespan assay, then grown on NGM plates covered with lawns of OP50 or OP50 (S+) until the late larval stage, L4 (3 days). Worms were then transferred to fresh plates, one animal per plate, with N = 8 animals per group. Thereafter, animals were transferred every 24 h to fresh plates with OP50 or OP50 (S+) bacteria until the end of the reproductive period77. The total number of progenies that grew up from each animal was counted, and the number of progenies for each concentration were averaged.
Stress resistance assays
Worms were grown on NGM plates containing OP50 or OP50 (S+) bacteria for a long time, and then subjected to stress resistance assays. Stress resistance assay were performed as previous described34. For oxidative stress, the 7-day old worms were transferred to a 24-well plate containing 30 mM paraquat (Sigma-Aldrich)33. The animals were monitored every 12 h and were scored as dead when they did not move in the liquid medium. These assays were each performed in three independent trials. Each group included at least 30 animals. For heat shock assays, the worms were transferred to an incubator set to 35 °C for 6 h and then scored for viability every hour33. Similarly, for the heavy metal stress assay, 3-day-old worms were transferred to a 24-well plate containing 10 mM K2Cr2O734. The animals were monitored every 12 h and were scored as dead when they did not move in the liquid medium. For the ultraviolet (UV) irradiation assay, 3-day-old animals were transferred to bacteria-free NGM plates and UV-irradiated at 600 J/m2 with an instrument equipped with a 5-watt 180-nm UV light bulb34. After UV irradiation, animals were transferred to fresh NGM plates with OP50 or OP50 (S+) and monitored daily for survival by gentle prodding with a platinum wire.
For stress resistance assays of animals fed different antioxidants, worms were grown from the age of 3 days on NGM plates containing antioxidants (DETP, vitamin C, or NAC) or on plates not containing antioxidants, and then subjected to stress resistance assays. For oxidative stress, 7-day old worms were transferred to a 24-well plate containing 30 mM paraquat (Sigma-Aldrich)33. The animals were monitored every 12 h and were scored as dead when they did not move in the liquid medium. These assays were each performed in three independent trials. Each group included at least 30 animals. Similarly, for the heavy metal stress assay, 3-day-old worms were transferred to a 24-well plate containing 10 mM K2Cr2O7. The animals were monitored every 12 h and were scored as dead when they did not move in the liquid medium.
RNA sequencing and data analysis
Age-synchronized nematodes were prepared as described above. Worms were grown on S medium with OP50 or OP50 (S+) bacteria. Three biological replicates of animals at 4 days and 12 days after the L4 stage were used for RNA sequencing. A total of 3 μg RNA per sample was used as input material for the RNA sample preparations. RNA quality was evaluated on a Nano Photometer spectrophotometer (IMPLEN, Munich, Germany), and the RNA integrity characterized using a Bioanalyzer 2100 (Agilent, Santa Clara, CA). Sequencing libraries were prepared following the protocol of the NEBNext Ultra RNA library Prep Kit (NEB) and sequenced on an Illumina HiSeq X Ten platform in the paired‐end mode (2 × 150 bp) through the service provided by Bionova. RNA-seq reads were aligned to the C. elegans reference genome (ws235) using HISAT2 (v2.0.578) with default parameters. Gene‐level read counts were calculated using HTSeq2 (v0.6.1p179) based on the Ensemble gene annotation v85. DEseq2 (v1.18.180) was used for data normalization.
Statistical analysis of differential expression was performed using the nbinomWaldTest in the DESeq2 package. Genes with an adjusted p‐value < 0.01 and |log2Foldchange| > 1 were defined as differentially expressed genes (DEGs).
Gene quantification through real-time PCR
Age-synchronized nematodes were prepared as described above. Worms were grown on S medium with OP50 and OP50 (S+) bacteria. Three biological replicates of animals at 4 days and 12 days after the L4 stage were used for the RNA isolation. The first-strand cDNA synthesis was then performed from the total RNA with the Maxima H Minus M-MuLV reverse transcriptase kit (Sangon Biotech, China). Expression levels of genes were measured with internal control of bmp-3. Real-time PCR was carried out by the 2 × SG Fast qPCR Master Mix Kit (Sangon Biotech, China) on Analytikjena & qTOWER3G touch (Analytikjena, Germany). Three independent trials were performed, and the relative mRNA quantification of the desired genes was analyzed by the comparative CT (ΔΔ CT) method46. The qRT-PCR primers were listed in Supplementary Table 20.
Statistics and reproducibility
All experiments were performed at least three times. For in vivo experiments in C. elegans, All the results were expressed as (mean value ± standard deviation). The statistical significance was calculated with the Student’s t test and log-rank test using GraphPad Prism. p values < 0.05 were considered to be significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
RNAseq data that support the findings of this study have been deposited in the Sequence Read Archive (SRA) at NCBI with the accession codes SRR16378842, SRR16378843, SRR16378844, and SRR16378845 (https://www.ncbi.nlm.nih.gov/sra/PRJNA772010). The authors declare that the other data supporting the findings of this study are available within the paper and its supplementary information files. Source data for figures are provided with the paper as Supplementary Data 1–8. The raw figure data that supporting Supplementary Figs. 1–3 are available in FIGSHARE with the identifiers https://doi.org/10.6084/m9.figshare.16909399.v1 (all the panels in Supplementary Fig. 1), https://doi.org/10.6084/m9.figshare.16909417.v1 (all the panels in Supplementary Fig. 2), https://doi.org/10.6084/m9.figshare.16909426.v1 (all the panels in Supplementary Fig. 3).
Change history
14 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s42003-021-02932-2
References
Tong, T. et al. Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proc. Natl Acad. Sci. USA 115, E2988–e2996 (2018).
Wang, L. et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Biol. 3, 709–710 (2007).
Zhang, Y. C. et al. Theoretical study on steric effects of DNA phosphorothioation: B-helical destabilization in Rp-phosphorothioated DNA. J. Phys. Chem. B 116, 10639–10648 (2012).
Wang, L. et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc. Natl Acad. Sci. USA 108, 2963–2968 (2011).
Cao, B. et al. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat. Commun. 5, 3951 (2014).
Chen, L. et al. Theoretical study on the relationship between Rp-phosphorothioation and base-step in S-DNA: based on energetic and structural analysis. J. Phys. Chem. B 119, 474–481 (2015).
Liu, G. et al. Structural basis for the recognition of sulfur in phosphorothioated DNA. Nat. Commun. 9, 4689 (2018).
Xiong, L. et al. A new type of DNA phosphorothioation-based antiviral system in archaea. Nat. Commun. 10, 1688 (2019).
Yu, H. et al. DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses. Nucleic Acids Res. 48, 8755–8766 (2020).
Xie, X. et al. Phosphorothioate DNA as an antioxidant in bacteria. Nucleic Acids Res. 40, 9115–9124 (2012).
Wu, T. et al. Mechanistic investigation on ROS resistance of phosphorothioated DNA. Sci. Rep. 7, 42823 (2017).
Huang, Q. et al. Defense mechanism of phosphorothioated DNA under peroxynitrite-mediated oxidative stress. ACS Chem. Biol. 15, 2558–2567 (2020).
Xu, T. et al. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 38, 7133–7141 (2010).
Xiong, X. et al. SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat. Microbiol. 5, 917–928 (2020).
Ellison, C. K. et al. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 3, 773–780 (2018).
Wu, Y. et al. RNA phosphorothioate modification in prokaryotes and eukaryotes. ACS Chem. Biol. 15, 1301–1305 (2020).
Sun, Y. et al. DNA phosphorothioate modifications are widely distributed in the human microbiome. Biomolecules 10, 1175 (2020).
Dai, D. et al. DNA phosphorothioate modification plays a role in peroxides resistance in Streptomyces lividans. Front. Microbiol. 7, 1380 (2016).
Kellner, S. et al. Oxidation of phosphorothioate DNA modifications leads to lethal genomic instability. Nat. Chem. Biol. 13, 888–894 (2017).
Yang, Y. et al. DNA backbone sulfur-modification expands microbial growth range under multiple stresses by its anti-oxidation function. Sci. Rep. 7, 3516 (2017).
Eruslanov, E. et al. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594, 57–72 (2010).
Armstrong, D. Preface. Advanced protocols in oxidative stress III. Methods Mol. Biol. 1208, v–vi (2015).
Büchter, C. et al. Myricetin-mediated lifespan extension in Caenorhabditis elegans is modulated by DAF-16. Int J. Mol. Sci. 14, 11895–11914 (2013).
Ray, A. et al. Mitochondrial dysfunction, oxidative stress, and neurodegeneration elicited by a bacterial metabolite in a C. elegans Parkinson’s model. Cell Death Dis. 5, e984 (2014).
Lee, S. J. et al. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136 (2010).
Wen, Y. et al. A highly sensitive ratiometric fluorescent probe for the detection of cytoplasmic and nuclear hydrogen peroxide. Anal. Chem. 86, 9970–9976 (2014).
Zhou, Z. et al. A dual-signal colorimetric and near-infrared fluorescence probe for the detection of exogenous and endogenous hydrogen peroxide in living cells. Sens. Actuators B 280, 120–128 (2019).
Gusarov, I. et al. Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152, 818–830 (2013).
Pincus, Z. et al. Autofluorescence as a measure of senescence in C. elegans: look to red, not blue or green. Aging 8, 889–898 (2016).
König, J. et al. Mitochondrial contribution to lipofuscin formation. Redox Biol. 11, 673–681 (2017).
Chen, Y. L. et al. Adiponectin receptor PAQR-2 signaling senses low temperature to promote C. elegans longevity by regulating autophagy. Nat. Commun. 10, 2602 (2019).
Komura, T. et al. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 14, 73–87 (2013).
Kato, M., Hamazaki, Y., Sun, S., Nishikawa, Y. & Kage-Nakadai, E. Clostridium butyricum MIYAIRI 588 increases the lifespan and multiple-stress resistance of Caenorhabditis elegans. Nutrients 10, 1921 (2018).
Park, H. H. et al. Survival assays using Caenorhabditis elegans. Mol. Cells 40, 90–99 (2017).
Reczek, C. R. et al. A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol. 13, 1274–1279 (2017).
Muhlemann, J. K. et al. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc. Natl. Acad. Sci. USA 115, E11188–e11197 (2018).
Kammeyer, A. et al. Oxidation events and skin aging. Ageing Res Rev. 21, 16–29 (2015).
Li, S. T. et al. DAF-16 stabilizes the aging transcriptome and is activated in mid-aged Caenorhabditis elegans to cope with internal stress. Aging Cell 18, e12896 (2019).
Gomez-Amaro, R. L. et al. Measuring food intake and nutrient absorption in Caenorhabditis elegans. Genetics 200, 443–454 (2015).
Wilson, M. A. et al. Blueberry polyphenols increase lifespan and thermotolerance in Caenorhabditis elegans. Aging Cell 5, 59–68 (2006).
Tharyan, R. G. Transcription factor nfyb-1 regulates mitochondrial function and promotes longevity induced by mitochondrial impairment. PhD thesis, Universität zu Köln (2019).
Gao, A. W. et al. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 113, 128–140 (2018).
Halaschek-Wiener, J. et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 15, 603–615 (2005).
Wan, Q. L. et al. Hypotaurine promotes longevity and stress tolerance via the stress response factors DAF-16/FOXO and SKN-1/NRF2 in Caenorhabditis elegans. Food Funct. 11, 347–357 (2020).
Lin, C. et al. Antistress and anti-aging activities of Caenorhabditis elegans were enhanced by Momordica saponin extract. Eur. J. Nutr. 60, 1819–1832 (2021).
Zhang, L. et al. Significant longevity-extending effects of EGCG on Caenorhabditis elegans under stress. Free Radic. Biol. Med. 46, 414–421 (2009).
Yang, Z. Z. et al. Lonicera japonica extends lifespan and healthspan in Caenorhabditis elegans. Free Radic. Biol. Med. 129, 310–322 (2018).
Jayarathne, S. et al. Tart cherry increases lifespan in Caenorhabditis elegans by altering metabolic signaling pathways. Nutrients 12, 1482 (2020).
Hang, W. et al. Significant longevity-extending effects of a tetrapeptide from maize on Caenorhabditis elegans under stress. Food Chem. 130, 254–260 (2012).
Vijg, J. et al. Genome instability and aging. Annu. Rev. Physiol. 75, 645–668 (2013).
Burhans, W. C. et al. DNA replication stress, genome instability and aging. Nucleic Acids Res 35, 7545–7556 (2007).
Minocherhomji, S. et al. Replication stress activates DNA repair synthesis in mitosis. Nature 528, 286–290 (2015).
Bae, Y. S. et al. Regulation of reactive oxygen species generation in cell signaling. Mol. Cells 32, 491–509 (2011).
Bertero, E. et al. Calcium signaling and reactive oxygen species in mitochondria. Circ. Res. 122, 1460–1478 (2018).
Nickel, A. et al. Mitochondrial reactive oxygen species production and elimination. J. Mol. Cell Cardiol. 73, 26–33 (2014).
Hou, Y. et al. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019).
Niccoli, T. et al. Ageing as a risk factor for disease. Curr. Biol. 22, R741–R752 (2012).
Zorov, D. B. et al. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 (2014).
Podmore, I. D. et al. Vitamin C exhibits pro-oxidant properties. Nature 392, 559 (1998).
Rietjens, I. M. et al. The pro-oxidant chemistry of the natural antioxidants vitamin C, vitamin E, carotenoids and flavonoids. Environ. Toxicol. Pharmacol. 11, 321–333 (2002).
Kaźmierczak-Barańska, J. et al. Two Faces of Vitamin C-Antioxidative and Pro-Oxidative Agent. Nutrients 12, 1501 (2020).
Carocho, M. et al. A review on antioxidants, prooxidants and related controversy: natural and synthetic compounds, screening and analysis methodologies and future perspectives. Food Chem. Toxicol. 51, 15–25 (2013).
Meydani, M. Nutrition interventions in aging and age-associated disease. Ann. N. Y. Acad. Sci. 928, 226–235 (2001).
Moreno-Macias, H. et al. Effects of antioxidant supplements and nutrients on patients with asthma and allergies. J. Allergy Clin. Immunol. 133, 1237–1244 (2014).
Polidori, M. C. Antioxidant micronutrients in the prevention of age-related diseases. J. Postgrad. Med. 49, 229–235 (2003).
Wu, Z. et al. Nutrients, microglia aging, and brain aging. Oxid. Med. Cell Longev. 2016, 7498528 (2016).
Jafarlou, M. et al. An overview of the history, applications, advantages, disadvantages and prospects of gene therapy. J. Biol. Regul. Homeost. Agents 30, 315–321 (2016).
Wirth, T. et al. History of gene therapy. Gene. 525, 162–169 (2013).
Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 24, 374–387 (2014).
Trapani, I. et al. Seeing the light after 25 years of retinal gene therapy. Trends Mol. Med. 24, 669–681 (2018).
Ma, C. C. et al. The approved gene therapy drugs worldwide: from 1998 to 2019. Biotechnol. Adv. 40, 107502 (2020).
Tepper, R. G. et al. PQM-1 complements DAF-16 as a key transcriptional regulator of daf-2-mediated development and longevity. Cell 154, 676–690 (2013).
Zečić, A. et al. DAF-16/FoxO in Caenorhabditis elegans and its role in metabolic remodeling. Cells. 9, 109 (2020).
Kimura, K. D. et al. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 (1997).
De Magalhaes Filho, C. D. et al. Visible light reduces C. elegans longevity. Nat. Commun. 9, 927 (2018).
Chen, J., et al. Metformin extends C. elegans lifespan through lysosomal pathway. Elife 6, e31268 (2017).
Dehghan, E. et al. Hydralazine induces stress resistance and extends C. elegans lifespan by activating the NRF2/SKN-1 signalling pathway. Nat. Commun. 8, 2223 (2017).
Kim, D. et al. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S. et al. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I. et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550 (2014).
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (31770070), and the National Key R&D Program of China (2018YFA0901200).
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.-L.Z. and Z.D. and T.Y.; investigation, H.Q., R.L., F.C., and D.W.; visualization, H.Q. and Y.-L.Z.; validation, Y.-L. Z. and H.Q.; funding acquisitions, Y.-L.Z.; writing-original draft preparation, H.Q.; writing-review and editing, Y.-L.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: George Inglis.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, Q., Li, R., Yi, T. et al. Phosphorothioate-DNA bacterial diet reduces the ROS levels in C. elegans while improving locomotion and longevity. Commun Biol 4, 1335 (2021). https://doi.org/10.1038/s42003-021-02863-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02863-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.