Abstract
Helical swimming in free-space is a common behavior among microorganisms, such as ciliates that are covered with thousands hair-like motile cilia, and is thought to be essential for cells to orient directly to an external stimulus. However, a direct quantification of their three-dimensional (3D) helical trajectories has not been reported, in part due to difficulty in tracking 3D swimming behavior of ciliates, especially Tetrahymena with a small, transparent cell body. Here, we conducted 3D tracking of fluorescent microbeads within a cell to directly visualize the helical swimming exhibited by Tetrahymena. Our technique showed that Tetrahymena swims along a right-handed helical path with right-handed rolling of its cell body. Using the Tetrahymena cell permeabilized with detergent treatment, we also observed that influx of Ca2+ into cilia changed the 3D-trajectory patterns of Tetrahymena swimming, indicating that the beating pattern of cilia is the determining factor in its swimming behavior.
Similar content being viewed by others
Introduction
Ciliates are ciliated protists that swim in the water along helical paths by beating the thousands of cilia coating the cell body1,2. Helical swimming, which is typical for a swimming cell at low Reynolds number2,3,4,5, is associated with its efficient movement for obtaining nutrients or for escaping from threats6,7,8,9,10. One such ciliate, Tetrahymena, has long been used as a model organism for a broad range of biological studies11. Its swimming pattern has been described to involve helical motions, as observed through classical microscopy, which is capable of imaging only one focal plane. However, Tetrahymena is a 3D entity, whose helical swimming trajectory is not limited to only one focal plane. In fact, the handedness of helical swimming trajectories of Tetrahymena has not been determined quantitatively, owing to the difficulty in measuring fast 3D motions (swimming velocity of approximately 500 µm s−1)12 of small (approximately 50 µm in length) and transparent cell bodies13. The trajectory of the swimming cell appears to different observers to be following either a right- or left-handed helical path due to an optical illusion, sometimes referred to as the “spinning dancer14,15” caused by a lack of spatial visual cues16. Another ciliate, Paramecium, which is approximately 5 times larger than Tetrahymena, has been reported to swim along left-handed helical paths1,17, and only two species of Paramecium, P. calkinsi, and P. duboscqui, have been reported to be right-handed helical swimmers, as observed under a classical microscope18,19. A precise determination of the 3D parameters of helical swimming motions of ciliates has not been conducted till date. 3D quantification of their trajectories would be very helpful for probing the swimming mechanism of ciliates.
Here we observed the 3D swimming behavior of ciliates Tetrahymena in free-space by 3D tracking of fluorescent microbeads phagocytosed by the cells. The 3D tracking was achieved using a three-dimensional prismatic optical tracking (termed tPOT) microscope20, which yields 3D positional information. We report that ciliate Tetrahymena thermophila swims along right-handed helical paths, while the cell body rotates in a right-handed manner along its longitudinal axis. Furthermore, Tetrahymena cells permeabilized with detergent treatment, which swim backward with Ca2+ stimulus21,22, showed various helical swimming patterns, including backward swimming along right-handed helical paths, forward swimming along left-handed helical paths, or frequent changes in these two behaviors in response to Ca2+ stimulus. Based on our study outcomes and available scientific literature, here, we discuss the possible mechanism underlying the formation of various helical swimming patterns formed by Tetrahymena in free-space.
Results and discussion
Direct observation of three-dimensional swimming trajectories of ciliate Tetrahymena in free-space
To determine the handedness of helical swimming of ciliate Tetrahymena in free-space (Supplementary Movie 1), 3D swimming trajectories of Tetrahymena cells were tracked using a tPOT microscope, originally developed by us for studying molecular motor proteins using quantum dots20, single fluorescent molecules23, and microbeads24. We applied tPOT microscopy to a ciliate swimming assay by tracking fluorescent microbeads (200 nm in diameter) inside the cells after their intake by phagocytosis (Fig. 1a–c and Supplementary Movie 2). Using this method with high-spatial-resolution (Supplementary Fig. 1), we found Tetrahymena thermophila to have right-handed helical swimming motions (Fig. 1d). Trajectories on x–y and x–z planes indicated sinusoidal oscillations, the phase of oscillation on the x–y plane being faster by approximately 90° than that on the x–z plane (Fig. 1e), confirming right-handed helical motion (Fig. 1f). T. thermophila always swims along right-handed helical paths (n = 60), whereas some cells swim straight or move randomly only after hitting the surface of a cover glass. An average helical pitch was estimated to be 129 ± 29 µm (mean ± standard deviation, SD) (Supplementary Fig. 2), which is robust to changes in the forward velocity (Supplementary Fig. 3). We also examined handedness of helical motions driven by Tetrahymena pyriformis (n = 100). Although T. thermophila and T. pyriformis are close evolutionary relatives among ciliates, they are fairly distant within species comprising the T. pyriformis complex25. Using the tPOT system, we found the helical motion with an average helical pitch of 132 ± 43 µm (mean ± SD) driven by T. pyriformis to show the same handedness as that observed for T. thermophila (Supplementary Fig. 4). Although forward and revolving velocities of T. pyriformis were higher than those of T. thermophila, helical pitches of the two were not significantly different (p = 0.79, Wilcoxson rank-sum test) (Supplementary Figs. 5 and 6), indicating that a right-handed helical swimming trajectory is surprisingly robust amongst related species. Moreover, we found other ciliates, Paramecium multimicronucleatum and Paramecium calkinsi, to show left- and right-handed helical trajectories (Supplementary Fig. 7), respectively, consistent with the previous report based on the observation under a classical optical microscope1,17,18 (Supplementary Movie 3).
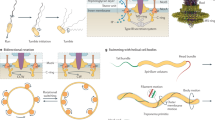
a Diagram of a tPOT microscope (not to scale). b Diagram of Tetrahymena swimming after ingestion of a fluorescent bead (not to scale). c Sequential images of the bead in a swimming cell, observed using a tPOT microscope. The filled and open arrowheads indicate the images (split by the prism) of the bead in a moving cell, respectively (time in seconds). d 3D plot of the bead (red) in a cell revealed right-handed helical swimming of T. thermophila. Dotted lines show the data acquired at 89 frames s−1, whereas the solid lines show the data averaged over every six frames. The arrow indicates the direction and distance of swimming during 100 ms along the x-axis. The x–y (pink) and x–z (blue) trajectories (e) and the y–z trajectory (f) of the bead in the cell in (d). Axes are rotated so that x-axis is parallel to the swimming direction. The trajectory of the first revolution is shown by the dotted black line and begins at the red open square (f). Based on this analysis, the handedness of helical swimming (red arrow) was checked. Forward velocity, revolving velocity, and helical pitch were 350 µm s−1, 24 rad s−1, and 92 µm, respectively. g Trajectories of two separate light spots in the same cell. The red and blue lines show the trajectories of the light spots, which were located posteriorly and anteriorly in the cell, respectively. h Relative trajectory of the anterior light spot when viewed from the posterior light spot (fixed at the origin).
We next explored whether the cell rotates along its longitudinal axis (i.e., roll) during right-handed helical swimming, and if so, which direction it rotates in (i.e., rolling direction). Since the 3D trajectory of one point in the cell body represented the sum of helical swimming motion and cell body rolling, our method was applied to track two separate light spots in single T. thermophila cell, corresponding to fluorescent beads in separate food vacuoles, which is able to extract the cell body rolling via the relative position of two spots (Supplementary Fig. 8). We found that the two spots in a single cell showed right-handed helices with a constant phase difference; the position of the front spot with respect to the rear spot showed right-handed rotation over time, indicating that the cell body rolled in a right-handed manner while swimming forward (Fig. 1g, h). Thus, the rolling direction coincided with the handedness of the helical swimming trajectories as predicted in previous theoretical reports2,3,26 and observed in other protists; Paramecium27,28, Chlamydomonas reinhardtii9,29 and Euglena gracilis30.
Three-dimensional observation of various swimming behaviors exhibited by Tetrahymena cells in response to depolarizing stimulation with Ca2+
In order to investigate the mechanism underlying the unidirectional helical swimming driven by Tetrahymena, we examined the 3D trajectories of Tetrahymena-driven backward swimming induced by calcium stimulation. An increase in intraciliary Ca2+ in T. thermophila permeabilized with detergent treatment has been known to trigger backward swimming along the longitudinal cell axis21,22 (hereafter, permeabilized cells with excess Ca2+ are referred to as “doping cells”). On tracking the 3D trajectories of doping cells in the presence of 100 µM CaCl2, we found that the backward swimming trajectories formed right-handed helices (Fig. 2a, blue), the handedness being same as that observed in forward swimming of non-doping cells (Supplementary Fig. 9 and Supplementary Movie 4). After adapting to the Ca2+ stimulus (within approximately 5–30 s), the cells stopped helical swimming, along with rotation on spot for a few seconds (Fig. 2a, light-blue), and regained forward right-handed helical swimming (Fig. 2a, green to red), as noted previously11. In addition to the backward right-handed helical swimming after the stimulation, we observed the slow forward left-handed helical swimming of doping cells (Fig. 2b, blue to light-blue) before adaptation. Some doping cells switched back and forth between the two different motions (Supplementary Fig. 9 and Supplementary Movie 5) before adaptation. Thus, the patterns of helical swimming motion, driven by individual cells, changed in response to Ca2+ stimulation.
a An example of a 3D swimming trajectory of T. thermophila immediately after depolarizing stimulation with Ca2+. The images represent the orientations of the cell body, and the blue and red arrows indicate the swimming directions. The colored line shows the data acquired at 89 frames s−1. The cell swam backward along a right-handed helical path, and then rotated on the spot. After adaptation, the cell swam forward along a right-handed helical path. b Another example of 3D swimming trajectory with stimuli. The cell swam forward along a left-handed helical path for several seconds and rotated on the spot. Then, it swam forward along a right-handed helical path. c Bidirectional swimming model. The relationship between the effective stroke direction of cilia and the handedness of helical swimming of the cell is overviewed. Normally, Tetrahymena swam forward and along a right-handed path. However, when the effective stroke direction changed, their swimming behavior also altered, usually recovering within 30 s.
As explained by Purcell, helical swimming trajectories are common among microorganisms moving at low Reynolds number4. For ciliate/flagellate protists, the motion of the cell at low Reynolds number could be driven by periodical whip-like beatings of cilia or flagella protruding from the cell body. The beat cycle consists of an effective stroke and a recovery stroke, which show distinct shapes31 and force profiles32. Each beat causes a discrete translation followed by a rotation of the cell body in free-space, and the repeated translation and rotation in three-dimension induces a helical swimming trajectory of the cell6,7,8,9,30, which has been generalized as the Helix Theorem30,33,34. Our direct 3D observation of the swimming behavior of the ciliate Tetrahymena showed that cells swam along right-handed helical paths with right-handed rolling (Fig. 1). This is probably due to the effective stroke direction of the cilia, taking place from the right anterior direction to the left posterior direction1. The repeated effective stroke simultaneously produces the forward movement and rotation of the cell, as the forward swimming velocity and revolving velocity are tightly coupled (Supplementary Fig. 3a). Although our current system failed to detect the effect of periodic ciliary beating (Supplementary Fig. 10), a recent experiment with flagellate Euglena gracilis has observed the effect of periodic flagellar beating30. Using a method to observe the changes in the 3D shapes of cilia at a high spatiotemporal resolution, the effect of periodic ciliary beating on helical swimming motion would be detected.
Our 3D tracking technique also revealed that the swimming behavior could be regulated by intraciliary Ca2+ perturbation (Fig. 2a, b). Since the mechanism of forward right-handed helical swimming remains unclear in case of the ciliate Tetrahymena, the mechanism by which depolarizing stimuli modulate chirality of the direction and helicity of swimming is also unclear. One possible explanation is as follows: the route of effective stroke can change, which determines not only forward/backward motion but also rolling direction (Fig. 2c)1,26. The repeated ciliary effective strokes towards the organism’s rear left would cause right-handed helical forward swimming, as mentioned above. For backward swimming, the strokes would be opposite, as the handedness of the helix is the same as in forward swimming (Fig. 2a). In fact, a reversal of the beating direction of cilia has been observed to revert the swimming direction of Paramecium35. In the present study, after stimulation by Ca2+, forward left-handed helical swimming (Fig. 2b) and frequent changes between right-handed backward and left-handed forward swimming (Supplementary Fig. 9 and Supplementary Movie 5) were also observed unexpectedly. This suggested that the handedness of helical swimming is not decided by the cell shape alone. Instead, a changeable route of the effective stroke is the key determinant of swimming behavior. The 3D orbits of ciliary strokes can change after Ca2+ stimulation21,36, and left-handed forward swimming is interpreted as an intermediate state of right-handed forward and backward swimming. Models addressing the mechanism by which an individual cilium produces directed force will be tested in the near future with further 3D studies combined with computational analysis.
Methods
Cell culture
Strain SB 255 of T. thermophila and strain W of T. pyriformis were cultured in 50 ml of SPP medium [1% Proteose Peptone No. 3, 0.2% glucose, 0.1% yeast extract, 0.003% Fe-EDTA, and 1% antibiotic antimycotic solution] (and 0.5% paromomycin, only for T. thermophila) in 125 ml bottles in an incubator at 15 °C. Serial transfers of the cells were performed once in every two weeks. The cells were transferred to another incubator at 30 °C the day before the observation.
Experimental setting
Before observation, 500 µl of culture medium containing cells was centrifuged at 400 × g for 1 min at ~25 °C, and 450 µl supernatant was discarded. Then, 5 µl of 0.2 µm fluorescent beads solution, 0.01% solids (F8810, Thermo Fisher Scientific) was added. The fluorescent beads are thought to be orally phagocytosed into food vacuoles at the base of the oral apparatus and further transported into the cell37. After 5 min, 950 µl of M-buffer [10 mM tricine, 0.5 mM MOPS, 50 μM CaCl2, 8 mM NaCl, pH 7.4] were added and the mixture was centrifuged at 400 × g for 1 min at ~25 °C. Next, 850 µl of the supernatant was discarded, and the cells were resuspended in 850 µl of new M-buffer. The suspension was centrifuged at 400 × g for 1 min at ~25 °C, and the steps were repeated five times. Finally, 950 µl of supernatant was discarded and after a five min incubation, the mixture containing cells was loaded into an observation chamber. The observation chamber consisted of two cover glasses (NEO Micro cover glass; Thickness No. 1; bottom: 24 × 36 mm, top: 18 × 18 mm, Matsunami, Tokyo, Japan) and four layers of double-sided tape. The height of the chamber was approximately 400 µm, which was much larger than the size of Tetrahymena cells. To prevent cell adsorption, the chamber was filled with 20 µl of 5 mg ml−1 Bovine serum albumin (Sigma-Aldrich) for 5 min and finally washed with 100 µl of M-buffer before use. Assays were carried out at 22.5 ± 1 °C.
Doping cell
The 0.2 µm fluorescent beads were ingested by the cells, as described in the “Experimental setting” section. For the preparation and observation of doping cells, KCl-buffer [50 mM KCl, 10 mM Tris-maleate (pH 7.0), 0.5% polyethylene glycol] was used instead of M-buffer. To make a hole in the cell membrane and allow Ca2+ to flow into the cells, 50 µl of M-Triton-buffer [10 mM tricine, 0.5 mM MOPS, 100 μM CaCl2, 8 mM NaCl, 0.0167% Triton-X, pH 7.4] were poured onto 5 µl of the mixture containing cells on the cover glass (NEO Micro cover glass; Thickness No. 1; 24 × 36 mm, Matsunami, Tokyo, Japan). Observations were performed within 30 s after the addition of M-Triton-buffer. In this study, the swimming trajectories of 16 doping cells were obtained; three cells swam backward along right-handed helical paths, six cells swam forward along left-handed helical paths, five cells swam switching continuously between the two swimming patterns, and two cells swam forward along right-handed helical paths.
3D prismatic optical tracking microscope
The swimming cells were observed under the 3D prismatic optical tracking (tPOT) microscope20, which provides z-positional information from planar images with submicron accuracy. tPOT microscope uses a prism to split one image into two and calculates z-positional information of the sample from the difference in y-positions of the two images. As illustrated in Fig. 1a, the back-focal-plane (BFP) of the objective (UPLXAPO 10×, NA 0.4 or 4×, NA 0.16, Olympus, Tokyo, Japan) was focused outside the camera port of an inverted microscope (IX70, Olympus, Tokyo, Japan) with achromatic Lens-1 (combined focal length 170 mm) to make an equivalent BFP (eBFP). To split the image beam path at the eBFP, a custom-made wedge prism (94.8°, Natsume Kougaku, Nagano, Japan) coated with an antireflective layer was precisely located at the eBFP. The two split images of a sample were focused on the camera focal plane by achromatic Lens-2 (combined focal length 170 mm). Images were recorded by CMOS camera (C14440-20UP, Hamamatsu Photonics, Hamamatsu, Japan) via HSR software (Hamamatsu Photonics) on a Windows 10 PC at 10.00 or 11.21 ms per frame (2 × 2 binning). For calibration of z-axis position, a custom-built stable stage (Chuukousha Seisakujo, Tokyo, Japan) equipped with a pulse motor (SGSP-13ACTR-BO, Sigma Koki, Tokyo, Japan) and controller (QT-CM2, Chuo Precision Industrial, Tokyo, Japan) was used to move the objective vertically while observing the stable fluorescent beads (0.2 µm or 0.5 µm in diameter, F8812, Thermo Fisher Scientific) placed inside the observation chamber filled with 0.2% agarose gel (Agarose S, Nippon Gene, Tokyo, Japan) or attached to the bottom of the chamber filled with M-buffer. The calculated z-position and actual z-position (as defined by the pulse motor) corresponded linearly over a range of ±50 µm from the focal plane, and there was no significant difference in the results of calibration by these methods.
Parameter calculation
The forward and revolving velocities were calculated by fitting the x displacement−time plots (averaged over every six frames) and revolution−time plots (averaged over every six frames) with a linear function using MATLAB. Helical pitches were calculated as forward velocities divided by the revolving velocities. Individual data are shown in Supplementary Figs. 2 and 5.
Statistics and reproducibility
Statistical analysis of data was done using MATLAB. Wilcoxon rank-sum test were used to compare the forward velocities, revolving velocities, and helical pitches, respectively, of T. thermophila and T. pyriformis (Supplementary Fig. 6). Tracking data for beads within the swimming cell were obtained from at least three independent experiments, and sample sizes are indicated in detail in the text or the “Methods” section.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All samples used in this study are available from the corresponding authors on request. The source data for graphs in the main figures are provided as Supplementary Data 1.
Code availability
Custom scripts were written for tracking beads within swimming cells with Igor Pro 5.05A, and for analysis of trajectories with MATLAB R2021a. These scripts are available on reasonable request.
References
Machemer, H. Ciliary activity and the origin of metachrony in Paramecium: effects of increased viscosity. J. Exp. Biol. 57, 239–259 (1972).
Blake, J. R. & Sleigh, M. A. Mechanics of ciliary locomotion. Biol. Rev. 49, 85–125 (1974).
Jennings, H. S. On the significance of the spiral swimming of organisms. Am. Nat. 35, 369–378 (1901).
Purcell, E. M. Life at low Reynolds number. Am. J. Phys. 45, 3–11 (1977).
Friedrich, B. M. & Jülicher, F. Steering chiral swimmers along noisy helical paths. Phys. Rev. Lett. 103, 1–4 (2009).
Crenshaw, H. C. Orientation by helical motion—I. Kinematics of the helical motion of organisms with up to six degrees of freedom. Bull. Math. Biol. 55, 197–212 (1993).
Crenshaw, H. C. & Edelstein-keshet, L. Orientation by helical motion—II. Changing the direction of the axis of motion. Bull. Math. Biol. 55, 213–230 (1993).
Crenshaw, H. C. Orientation by helical motion—III. Microorganisms can orient to stimuli by changing the direction to their rotational velocity. Bull. Math. Biol. 55, 231–255 (1993).
Crenshaw, H. C. A new look at locomotion in microorganisms: rotating and translating. Am. Zool. 36, 608–618 (1996).
Jikeli, J. F. et al. Sperm navigation along helical paths in 3D chemoattractant landscapes. Nat. Commun. 6, 1–10 (2015).
Hennessey, T. M. & Lampert, T. J. Behavioral bioassays and their uses in Tetrahymena. Methods Cell Biol. 109, 393–410 (2012).
Lampert, T. J., Coleman, K. D. & Hennessey, T. M. Chemoattraction to lysophosphatidic acid does not require a change in membrane potential in Tetrahymena thermophila. Cell Biol. Int. 35, 519–528 (2011).
Ohmura, T. et al. Simple mechanosense and response of cilia motion reveal the intrinsic habits of ciliates. Proc. Natl Acad. Sci. USA 115, 3231–3236 (2018).
Kayahara, N. Spinning Dancer. Procreo Flash Design Laboratory. http://www.procreo.jp/labo/silhouette.swf (2003).
Troje, N. F. & McAdam, M. The viewing-from-above bias and the silhouette illusion. Iperception 1, 143–148 (2010).
Wallach, H. & O’Connell, D. N. The kinetic depth effect. J. Exp. Psychol. 45, 205–217 (1953).
Mogami, Y. & Baba, S. A. Super-helix model: a physiological model for gravitaxis of Paramecium. Adv. Sp. Res. 21, 1291–1300 (1998).
Wichterman, R. The Biology of Paramecium (Springer US, 1986).
Shi, X., Jin, M. & Liu, G. Rediscovery of Paramecium duboscqui Chatton & Brachon, 1933, and a description of its characteristics. J. Eukaryot. Microbiol. 44, 134–141 (1997).
Yajima, J., Mizutani, K. & Nishizaka, T. A torque component present in mitotic kinesin Eg5 revealed by three-dimensional tracking. Nat. Struct. Mol. Biol. 15, 1119–1121 (2008).
Naitoh, Y. & Kaneko, H. Reactivated triton-extracted models off paramecium: modification of ciliary movement by calcium ions. Science 176, 523–524 (1972).
Izumi, A. & Miki‐Noumura, T. Tetrahymena cell model exhibiting Ca‐dependent behavior. Cell Motil. 5, 323–331 (1985).
Fujimura, S. et al. Dissection of the angle of single fluorophore attached to the nucleotide in corkscrewing microtubules. Biochem. Biophys. Res. Commun. 485, 614–620 (2017).
Maruyama, Y. et al. CYK4 relaxes the bias in the off-axis motion by MKLP1 kinesin-6. Commun. Biol. 4, 180 (2021).
Nanney, D. L., Park, C., Preparata, R. & Simon, E. M. Comparison of sequence differences in a variable 23S rRNA domain among sets of cryptic speacies of ciliated protozoa. J. Eukaryot. Microbiol. 45, 91–100 (1998).
Jung, I., Powers, T. R. & Valles, J. M. Evidence for two extremes of ciliary motor response in a single swimming microorganism. Biophys. J. 106, 106–113 (2014).
Jennings, H. S. Studies on reactions to stimuli in unicellular organisms. II.—The movements and motor reactions of Paramecium. Am. J. Physiol. 2, 311–341 (1899).
Crenshaw, H. C., Ciampaglio, C. N. & McHenry, M. Analysis of the three-dimensional trajectories of organisms: estimates of velocity, curvature, and torsion from positional information. J. Exp. Biol. 203, 961–982 (2000).
Kamiya, R. & Witman, G. B. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 98, 97–107 (1984).
Rossi, M., Cicconofri, G., Beran, A., Noselli, G. & DeSimone, A. Kinematics of flagellar swimming in Euglena gracilis: helical trajectories and flagellar shapes. Proc. Natl Acad. Sci. USA 114, 13085–13090 (2017).
Baba, S. A. Regular steps in bending cilia during the effective stroke. Nature 282, 717–720 (1979).
Katoh, T. A. et al. Three-dimensional tracking of microbeads attached to the tip of single isolated tracheal cilia beating under external load. Sci. Rep. 8, 1–9 (2018).
Shapere, A. & Wilczek, F. Geometry of self-propulsion at low Reynolds number. J. Fluid Mech. 198, 557–585 (1989).
Cicconofri, G. & DeSimone, A. Modelling biological and bio-inspired swimming at microscopic scales: recent results and perspectives. Comput. Fluids 179, 799–805 (2019).
Iwadate, Y. Photolysis of caged calcium in cilia induces ciliary reversal in Paramecium caudatum. J. Exp. Biol. 206, 1163–1170 (2003).
Eckert, R. Bioelectric control of ciliary activity. Science 176, 473–481 (1972).
Guerrier, S., Plattner, H., Richardson, E., Dacks, J. B. & Turkewitz, A. P. An evolutionary balance: conservation vs innovation in ciliate membrane trafficking. Traffic 18, 18–28 (2017).
Acknowledgements
We thank Kyohei Matsuda, Mitsuhiro Sugawa, and Yoko Y. Toyoshima for critical discussion. T. pyriformis was kindly gifted by Kentaro Nakano, University of Tsukuba, Japan. Paramecium strains were provided by NBRP Paramecium lab, Yamaguchi Univ. with support in part by MEXT National Bio-Resource Project. This work was supported in part by JSPS KAKENHI (grant numbers JP20K06635, JP21K19252 to J.Y.); MEXT KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (grant numbers JP19H05357, JP21H00386 to J.Y.).
Author information
Authors and Affiliations
Contributions
J.Y. designed the project. A.M. and M.Y. prepared biological samples. A.M. performed and analyzed behavioral bioassays. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks Antonio Desimone and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Quan-Xing Liu and Luke R. Grinham. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marumo, A., Yamagishi, M. & Yajima, J. Three-dimensional tracking of the ciliate Tetrahymena reveals the mechanism of ciliary stroke-driven helical swimming. Commun Biol 4, 1209 (2021). https://doi.org/10.1038/s42003-021-02756-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02756-0
This article is cited by
-
Membrane-bound myosin IC drives the chiral rotation of the gliding actin filament around its longitudinal axis
Scientific Reports (2023)
-
Torque generating properties of Tetrahymena ciliary three-headed outer-arm dynein
Scientific Reports (2022)
-
Controlling non-controllable scallops
Meccanica (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.