Abstract
The enpp ectonucleotidases regulate lipidic and purinergic signalling pathways by controlling the extracellular concentrations of purines and bioactive lipids. Although both pathways are key regulators of kidney physiology and linked to human renal pathologies, their roles during nephrogenesis remain poorly understood. We previously showed that the pronephros was a major site of enpp expression and now demonstrate an unsuspected role for the conserved vertebrate enpp4 protein during kidney formation in Xenopus. Enpp4 over-expression results in ectopic renal tissues and, on rare occasion, complete mini-duplication of the entire kidney. Enpp4 is required and sufficient for pronephric markers expression and regulates the expression of RA, Notch and Wnt pathway members. Enpp4 is a membrane protein that binds, without hydrolyzing, phosphatidylserine and its effects are mediated by the receptor s1pr5, although not via the generation of S1P. Finally, we propose a novel and non-catalytic mechanism by which lipidic signalling regulates nephrogenesis.
Similar content being viewed by others
Introduction
Vertebrate kidney organogenesis is orchestrated by numerous signalling pathways and transcription factors regulating the proliferation and differentiation of diverse cell types to form the functional kidney. Despite the differences in complexity and organization of the three vertebrate kidneys, pronephros, mesonephros and metanephros, there is a remarkable conservation of molecular mechanisms during their development1. The pronephros, the functional embryonic kidney in amphibians, is a simple, easily accessible organ, which displays structural similarities to the other more complex kidney forms. Therefore, it has become an ideal model system to study molecular regulation during nephrogenesis and renal pathologies2,3,4,5.
In mammals, lipidic and purinergic pathways regulate metanephric physiology and their deregulation has been linked to acute renal injury and chronic kidney diseases including renal fibrosis polycystic kidney disease, renal cell carcinoma, nephritis or diabetic nephropathy6,7,8,9,10. However, their potential roles during renal development have not been fully established, although the bioactive lipid sphingosine-1-phosphate (S1P) has been implicated during kidney branching11. Purines, mostly ATP and its derivatives, and bioactive lipids, S1P and lysophosphatidic acid (LPA), can function as extracellular ligands for G protein-coupled cell surface receptors12,13. Their availability for these receptors, in the extracellular space, is regulated by the activities of several membrane-bound enzymes, such as the ectonucleotidases, which are major regulators of renal health and disease10,14. The enpp (ectophosphodiesterase/nucleotide phosphohydrolase) proteins, which belong to the ectonucleotidase subfamily, are key regulators of both purinergic and lipidic signalling pathways with their dual enzymatic activities of hydrolysing purines and generating S1P and LPA bioactive lipids15. We have demonstrated that the pronephros is the major site of expression for the amphibian enpp genes family, in particular, enpp4 is highly expressed in Xenopus laevis pronephric tubules16. These data provided the first temporal and spatial embryonic expression profile for this evolutionally conserved enzyme which remains functionally poorly understood17,18,19. In the present study, we investigated the function of enpp4 during pronephric development.
We demonstrate that Enpp4 function is crucial during kidney formation. While its knock-down leads to kidney formation defects, the overexpression of wild-type Enpp4, but not an inactive enzymatic protein, induces the formation of ectopic pronephroi characterized mostly by the presence of proximal tubule markers but in rare occasion of more distal tubule markers. These effects are mediated by the lipidic receptor S1pr5 and we also show that Enpp4 specifically binds to phosphatidylserine, implying a role for bioactive lipids in pronephrogenesis. Finally, we provide evidence that enpp4 misexpression alters the expression of members of the Notch, Wnt and RA signalling pathways and we propose a model for the mechanisms of action for Enpp4 and lipidic signalling in kidney development.
Results
Overexpression of Enpp4 results in ectopic pronephric tubules formation
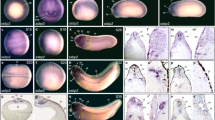
To analyse potential functional roles of Enpp4 during pronephros development, we first undertook a gain of function approach by performing immunostaining with pronephric tubules specific antibodies20 on stage 41 embryos (Fig. 1a–o, and Supplementary Table 1 for raw data and statistical analyses). Enpp4 overexpression altered proximal pronephric tubules formation, in nearly 50% of the analysed embryos and induced ectopic (23%) and enlarged (18%) regions of the 3G8 staining domain (n = 91, Fig. 1b–e; Supplementary Table 1). Distal tubules were less affected, with 31% of the analysed embryos displaying abnormal 4A6 staining. Ectopic 4A6 staining was rare (2%), with enlarged more distal tubule staining being the predominant phenotype (20%). Enpp4-induced phenotypes are significantly different compared to those of LacZ controls (3G8: p < 0.001 and 4A6: p < 0.05). Ectopic pronephroi were observed only when injections were performed into regions fated to become the lateral region of embryos (V2 blastomere) (Supplementary Fig. 1a). Enpp4 overexpressing embryos displaying ectopic 3G8 staining (n = 5) were analysed by transverse section. Eight of the nine ectopic tubules sectioned had epithelial tubule structure complete with a lumen (Fig. 1f–j), which were similar to normal pronephric tubule structure (Fig. 1k, l). Similar significant renal phenotypes were observed following mouse Enpp4 mRNA injection (p < 0.001, n = 63; Fig. 1m). However, no ectopic pronephric tubules were observed upon overexpression of Enpp4 mutant constructs containing a point mutation in the putative catalytic domain (T72A, T72S) or metal cation binding domain (D36N, D189N) (Fig. 1n, o)16. These data suggest that ectopic proximal pronephric tissues formation caused by Enpp4 overexpression depends on its catalytic activity.
a Schematic diagram of pronephric structural components showing the expression domain for each marker used in this study, adapted from ref. 21. G: glomus, PT: proximal tubule, IT: intermediate tubule, DT: distal tubule, CT: collecting tubule. b–y Embryos injected with 2 ng of enpp4 and 250 pg of LacZ mRNAs were examined by 3G8/4A6 antibody staining (b–o) or whole-mount in situ hybridization with the following probes: slc5a1.1 (p), slc12a1 (q), clcnkb (r) and gata3 (s) at stage 37/38; wt1 (t) and nphs1 (u) at stage 32; lhx1 (v, x) and pax8 (w, y) at stages 28 and 14. f–l Transverse sections of the embryo shown in panels (d) and (e) were cut in the anterior–posterior registers indicated by lines in panel (e). A higher magnification image (i) of ectopic pronephros in the somite indicated by square in (f) and of control kidney (k) and counterstained with Hoechst to indicate nuclei (j, l). Embryos injected with 2 ng of mouse wild-type Enpp4 (m), X. laevis mutated in the putative catalytic site (n) or in the cation binding site (o) and 250 pg of LacZ mRNAs were examined by 3G8/4A6 antibody staining. The asterisk denotes the uninjected side of each embryo. Arrowheads indicate ectopic marker staining. Blank arrowheads in (s) indicate the anterior limit of gata3 expression. See also Supplementary Table 1 for raw data and statistical analyses and Supplementary Fig. 1.
Overexpression of Enpp4 disturbs proximal-distal patterning of pronephros
To further investigate this phenotype, embryos injected with enpp4 mRNA were examined at stage 37 by whole-mount in situ hybridization using pronephric specific markers, slc5a1.1, slc12a1, clcnkb and gata3, which mark the various proximal/distal tubule segments21,22 (Fig. 1a and p–s, see also Supplementary Table 1). Interestingly, Enpp4 overexpressing embryos showed ectopic and enlarged staining of slc5a1.1 (proximal tubule marker, ectopic 30%, enlarged 14%; n = 57; p < 0.001) and slc12a1 (marker of intermediate tubules, ectopic 17%, enlarged 25%; n = 57, p < 0.001) domains (Fig. 1p–q). Injection of enpp4 mRNA failed to induce any separate ectopic clcnkb expression although the normal domain of expression (intermediate and distal tubule) was somewhat enlarged on the injected side (19%, n = 58, p < 0.01; Fig. 1r). The gata3 expression domain (distal and collecting tubules) was relatively normal, although its anterior limit of expression, determined relative to the somite number, was slightly more posterior in more than half of the injected embryos (58%, n = 43, p < 0.001; Fig. 1s). This might reflect a change in anterior/posterior patterning induced by Enpp4 overexpression.
Injection of enpp4 mRNA induced enlarged and reduced expression domains of both glomus marker wt1 and nphs1 at stage 33/34 but ectopic glomus staining was only observed in rare cases (Fig. 1t–u; Supplementary Table 1a). Although the statistical significance of these phenotypes was demonstrated (Supplementary Table 1b), we were not able to conclude on the exact Enpp4 effects on this structure.
Taken together, the results demonstrate that enpp4 mRNA injection altered pronephros formation, leading to enlarged expression domains of markers of the entire tubule segments and to ectopic pronephric structures containing mostly domains of proximal and, in rare occasions, distal tubules marker genes.
Overexpression of Enpp4 upregulates early kidney markers expression without altering mesoderm formation
Embryos injected with enpp4 mRNA were also examined by whole-mount in situ hybridization using early pronephros anlagen markers irx1, lhx1, pax823,24 and compared to lacZ mRNA injected embryos (see Supplementary Table 1). At stage 28, expression of both lhx1 (61%, n = 51) and pax8 (70%, n = 50) was significantly (p < 0.001) expanded especially in posterior parts of pronephric anlagen, with areas of intense staining consistent with that of the more anterior presumptive tubules (Fig. 1v–w). At early neurula stages (Fig. 1x–y), the expansion of pax8 (17%, n = 70) expression domain was also observed (Fig. 1y). Furthermore, ectopic pax8 expression was also induced following enpp4 RNA injection at both stages analysed (neurula stage, 21%; stage 28, 2%; p < 0.001; Supplementary Table 1b). Lhx1 expression was also altered at early neural stages, but no ectopic lhx1 expression was observed (Fig. 1x, Supplementary Table 1b). Furthermore, irx1 expression domain was not altered following Enpp4 overexpression (Supplementary Fig. 1b).
Since normal somite development is a prerequisite for pronephros development, enpp4 RNA injected embryos were analysed by whole-mount in situ hybridization using the muscle marker myh4 at stage 33/34. The expression pattern was normal in all injected embryos (n = 55; Supplementary Fig. 1c; Supplementary Table 1). Enpp4 overexpression did not also alter the expression of pan-mesoderm marker, xbra at stage 10.5 (n = 46; Supplementary Fig. 1d and Supplementary Table 1). Therefore, we conclude that, enpp4 mRNA injection had no gross effects on mesoderm induction per se or on somite development.
Morpholino knock-down of enpp4 results in smaller pronephros formation
To determine whether Enpp4 is required for normal pronephros development, we have undertaken a loss of function approach using two specific anti-sense morpholino oligonucleotides (MOs) (Fig. 2, Supplementary Fig. 2a–c for specificity and efficiency of the MOs and Supplementary Table 2). The overall morphology of the embryos appeared normal in enpp4 morphants. Moreover, the expression pattern of the somitic myh4, muscle myoD and early mesoderm xbra markers was unaffected in enpp4 MO1 injected embryos (myh4 100%, n = 47 and xbra 84%, n = 19; Supplementary Fig. 2d, e) suggesting that any kidney phenotypes observed in enpp4 morphants are not due to general mesoderm defects.
Embryos targeted with 10 ng of enpp4 MO1 or 10 ng of both enpp4 MOs (c, n) and 250 pg of lacZ mRNA were examined by 3G8/4A6 antibody staining (a–c) or whole-mount in situ hybridization with the following probes: slc5a1.1 (d), slc12a1 (e, f), clcnkb (g) and gata3 (h) at stage 37/38; wt1 (i) and nphs1 (j) at stage 33/34, lhx1 (k–n) at stages 28, 24 and 14, pax8 (o) at stage 14. The embryo shown in (f) was co-injected with 2 ng of mouse Enpp4 mRNA to rescue enpp4 knock-down phenotype. The asterisk denotes the control, uninjected side of each embryo. Blank arrowheads in (h) indicate the anterior limit of gata3 expression. See also Supplementary Table 2 for raw data and statistical analyses and Supplementary Figs. 2 and 3.
Injection of enpp4 MO1 resulted in a significant reduction of expression of both 3G8 (65%, p < 0.001) and 4A6 (28%, p < 0.05) (n = 107, Fig. 2a, b, Supplementary Table 2), indicating that a smaller pronephros had formed. A similar phenotype was observed following enpp4 MO2 injection (3G8 49%, 4A6 24%, n = 87; Supplementary Fig. 2f, g) and was worsened when both MO were injected together (Fig. 2c). These results indicate that Enpp4 is required for both proximal and distal pronephric tubule development.
Enpp4 knock-down in embryos showed a significant reduced expression of slc5a1.1 (58%, n = 64, p < 0.001) and slc12a1 (56%, n = 75, p < 0.01) with MO1 (Fig. 2d, e) as well as with MO2 (Supplementary Fig. 2h, i). Rescue experiments performed by co-injecting mouse Enpp4 mRNA (2 ng) with enpp4 MO1 or MO2 (10 ng each, n = 72 or 28, respectively) restore partially but significantly (p < 0.001) the normal phenotype of slc12a1 staining domain confirming the specificity of the knock-down of enpp4 expression on pronephric development (Fig. 2f and Supplementary Fig. 2j). Ectopic slc12a1 expression was also observed in some embryos (11% and 50% with MO1 or MO2, respectively) consistent with Enpp4 overexpression phenotype. The clcnkb expression in the intermediate tubules and gata3 anterior expression domain were reduced after enpp4 MO1 (53.5%, n = 40 and 31%, n = 42, respectively) but these differences are not statistically significant p > 0.05) (Fig. 2g, h). Enpp4 knock-down has no effect on glomus formation, as the expression of wt1 and nphs1 was normal in most of enpp4 MO1 injected embryos at stage 33/34 (wt1 = 94%, n = 34 and nphs1 = 83%, n = 41) (Fig. 2i, j). These results suggest that enpp4 knock-down affected pronephric tubule, especially proximal and intermediate segments, differentiation rather than just the proximal-distal patterning of pronephric tubule segmentation.
To address potential Enpp4 roles during early phases of pronephros development, we tested by RT-PCR its expression at key stages during kidney development in dissected developing pronephric tissues (Fig. 3a). At later stages, enpp4 expression profile is in agreement with our published in situ hybridization data16. However, weak expression is also detected in the embryonic kidney from stage 12.5 and is upregulated by stage 26. These data suggest that Enpp4 might be involved during early pronephric developmental phases. The expression domain of lhx1 and pax8 was altered following knock-down of enpp4 by MO injection (see Supplementary Table 2 for raw data and statistical analyses). At stage 28, expression of lhx1 was reduced especially in posterior elements of the pronephric anlagen (35%, n = 23; Fig. 2k), although its expression in presumptive proximal tubules was sometimes unaffected or expanded. At stage 24, the expression domain of lhx1 was clearly reduced (75%, n = 20, Fig. 2l), suggesting involvement of Enpp4 in early pronephros differentiation. Injection of enpp4 MO2 caused similar phenotypes at both stages (stage 28, 30%, n = 20; stage 24, 55%, n = 20; Supplementary Fig. 2k–l). At early neurula stages, expression of lhx1 was also reduced following enpp4 MO1 or MO2 injection (MO1 37%, n = 46; MO2 64%, n = 61; p < 0.001; see also Supplementary Table 2) and even absent after MO1 injection (MO1 31%, n = 46) (Fig. 2m and Supplementary Fig. 2m). The injection of both MO’s resulted in a stronger reduction of lhx1 expression (n = 31; Fig. 2n). A reduction of pax8 expression was also observed at the neurula stages following enpp4 MO1 or MO2 injection (Fig. 2o and Supplementary Fig. 2n).
Developing pronephric anlagen or pronephric tubules were dissected as indicated, from whole X. laevis embryos and total RNA extracted. RT-PCR was performed on pronephric dissected tissues and control whole embryos along with negative and linearity controls. a Comparative expression pattern of the enpp genes and pronephric and muscle marker genes controlling the quality of the dissections. b Comparative expression profile of the lpa receptors. c Comparative expression profile of the s1pr genes.
Taken together, the Enpp4 knock-down and rescue experiments demonstrate that normal levels of enpp4 expression are required for normal pronephric development.
Enpp4 misexpression phenotypes are distinct from those following enpp6 misexpression
To address if the ectonucleotidase Enpp6, also expressed in the proximal pronephric tubules16, can compensate for Enpp4 loss of function, we performed single or double enpp4/enpp6 knock-down and rescue experiments (Supplementary Fig. 3 and Supplementary Table 2). Enpp6 depletion induced the formation of a smaller pronephros on the injected side, in the similar frequency than enpp4 knock-down (Supplementary Fig. 3b, c). The co-injection of enpp4 MO2 and enpp6 MO resulted in the formation of reduced 3G8 (64%, n = 76) and 4A6 (62%, n = 76) positive tissues and was not statistically different from the effects of enpp4 MO2, demonstrating that the enpp6 MO did not worsen the renal phenotype caused by enpp4 knock-down (Supplementary Fig. 3a). Pronephric formation was altered following Enpp6 overexpression, with reduced or absent, but never ectopic, pronephros observed and injection of enpp6 mRNA did not rescue the enpp4 MO2 phenotype (Supplementary Fig. 3d, e). Taken together, these data suggest that there is no functional redundancy between Enpp4 and Enpp6 ectonucleotidases.
Enpp4 misexpression affects expression of several components of the RA, Notch and Wnt signalling pathways
Since retinoic acid (RA), Notch and Wnt signalling pathways are involved in pronephros formation and patterning25,26,27,28,29,30 and the timing of the endogenous expression of many components of these pathways overlap, we hypothesized that Enpp4 might affect these pathways. We therefore examined embryos injected with enpp4 mRNA (Fig. 4a–g) or enpp4 MO1 (Fig. 4h–n) by in situ hybridization for alterations in expression domain of representative members e.g. ligands (Dll1, Jag1, Wnt4) receptors (Notch1) and metabolic enzymes (Raldh1a2, Rdh10, Cyp26a1) of these three pathways (see also Supplementary Table 3).
a–g Embryos targeted with 2 ng of enpp4 and 250 pg of LacZ mRNA or h–n 10 ng of enpp4 MO1 and 250 pg of LacZ mRNA were fixed at stage 28 and examined by whole-mount in situ hybridization with the following probes: raldh1a2 (a, h), rdh10 (b, i), cyp26a1 (c, j), notch1 (d, k), dll1 (e, l), jag1 (f, m) and wnt4 (g, n). The asterisks denote the control, uninjected sides. Arrowheads indicate ectopic staining of the marker gene (see also Supplementary Table 3 and Supplementary Fig. 4).
Enpp4 overexpression induced ectopic and enlarged raldh1a2 and rdh10 expression domains in the pronephric region (p < 0.001, n = 37 and 34, respectively, Fig. 4a, b and Supplementary Table 3). Enpp4 knock-down reduced their expression in pronephric region of ~20% of analysed embryos but this phenotype is not significant (p > 0.05, n = 33 for each probe, Fig. 4h, i and Supplementary Table 3). Raldh1a2 expression was unaffected in the pharyngeal arches. In contrast, cyp26a1 expression was normal in the pronephric region following enpp4 mRNA or MO injection (98%, n = 44 and 88%, n = 42, p > 0.05, respectively) (Fig. 4c, j). Both enpp4 mRNA and MO injection disturbed rdh10 and cyp26a1 expression in the somites. These results suggest Enpp4 controls the expression of enzymes involved in RA synthesis and potentially might act upstream of RA signalling. To verify this, enpp4 expression, along with pax8 and lhx1, was analysed in animal caps. Enpp4 expression is not induced in animal caps treated with RA for 3 h (Supplementary Fig. 4). This confirms the epistatic relationship between Enpp4 and RA signalling.
Enpp4 overexpression revealed significant enlarged expression domains of notch1 (53% n = 75, p < 0.001, Fig. 4d). Notch1 expression was normal in the majority of embryos after enpp4 knock-down (80%, n = 41, Fig. 4k). Enpp4 mRNA injection caused ectopic (40%) and enlarged (23%) expression domains of dll1 (n = 81, p < 0.001 Fig. 4e), while MO-injected embryos showed dll1 reduced expression (33%, n = 46, Fig. 4l). Ectopic (20%) and enlarged (44%) jag1 expression domains were observed following Enpp4 overexpression (n = 81, p < 0.001, Fig. 4f), while MO1 injection reduced its expression domain (in 38% of the analysed embryos, n = 42, Fig. 4m). These results suggest that Enpp4 also regulates members of the Notch signalling pathway and that jag1 expression is more affected by enpp4 depletion than dll1 expression. Since Rnfg overexpression caused ectopic pronephroi formation28, we further addressed the link between Enpp4 and the Notch pathway by injecting enpp4 mRNA or MO2 in presence of rfng mRNA or MO. Our data show that modulation of notch-ligand interactions by Fringe proteins alters Enpp4 pronephric phenotypes, although differences are not significant (Supplementary Fig. 5 and Supplementary Table 3).
Finally, enpp4 mRNA injected embryos showed enlarged (32%) and ectopic (17%) wnt4 expression domains (n = 41, Fig. 4g) while expression of wnt4 was reduced in most of the enpp4 MO1 injected embryos (82%, n = 39, p < 0.001 Fig. 4n, Supplementary Table 3). These data suggest that Enpp4 is necessary and sufficient for promoting pronephric wnt4 expression.
Xenopus Enpp4 is localized to the plasma membrane
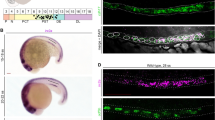
To address the cellular localization of the amphibian Enpp4 protein, we generated a specific polyclonal antibody against the full-length Xenopus protein (see Supplementary Table 4 for specificity evaluation of the antibody) and expressed Xenopus wild type (WT), T72S mutant and mouse Enpp4 cDNA in CHO cells by transient transfection. Xenopus Enpp4 WT protein was detected, by western blotting, in whole cells and in the membrane fractions, but not in the soluble fractions (Fig. 5a). Immunofluorescence experiments confirmed Enpp4 expression at the cell membrane (Fig. 5b). These results show that Xenopus Enpp4 is a transmembrane protein, as its mouse ortholog31. Unfortunately, we failed to detect the endogenous Enpp4 expression in Xenopus embryos using the anti-XlEnpp4 antibody.
a CHO cells were transfected with Xlenpp4-pcDNA3.1 or empty vector (control) and the cellular distribution of Enpp4 determined by western blotting using anti-XlEnpp4 antibody and proteins extracts from membrane (mb), soluble (sb) or whole cells (wh) fractions. b Representative images of the cellular distribution of Enpp4 determined by immunofluorescence using anti-XlEnpp4 or anti-mEnpp4 antibodies from CHO cells transfected with Xlenpp4-pcDNA3.1, Xlenpp4T72S-pcDNA3.1, mEnpp4-pcDNA3.1 or empty pcDNA3.1 vector (control). Corresponding brightfield images are also presented. Scale bar represents 20 µm.
Phospholipid receptors are expressed in the developing pronephros along with the enpp4 gene
Based on sequence homology of Xenopus Enpp family16, Enpp4 is more related to lipid-hydrolysing Enpp6 and 7 enzymes. We therefore hypothesized that the roles of transmembrane-bound Enpp4 during pronephrogenesis might be linked to the lipidic signalling pathway. To test if phospholipid receptors might mediate Enpp4 functions, we established the expression profiles of lpa and s1p receptor family members previously identified32 by RT-PCR in kidney dissected tissues (Fig. 3b, c). All lpa receptors, except lpar3 and 5, are expressed in pronephric tissues at a similar level from the time of kidney specification to late differentiation, confirming their ubiquitous expression profile during Xenopus embryogenesis32. The s1p receptors display different expression profiles, with s1pr5 being the only family member to be expressed in the developing kidney at every stage analysed, particularly in the presumptive pronephric tissue at stage 12.5. No such renal expression was detected by in situ hybridization in our previous study, although expression in marginal zone of blastula embryos was detected by RT-PCR32. These data suggest that the pronephric level of expression of these lipidic receptors, especially s1pr5, is relatively low, under the in situ hybridization detection level.
Overexpression of s1pr5 enhances Enpp4 function to induce ectopic pronephros
In order to identify whether a lipidic receptor is involved in Enpp4 phenotypes, s1pr and lpar overexpression analyses were carried out by injecting 2 ng of s1pr5, s1pr1, lpa1.1 and p2y10 mRNAs alone or in combination with 1 ng of enpp4 mRNA alone (Fig. 6a–d; Supplementary Fig. 6a–f; Supplementary Table 5) i.e half of the optimal dose to generate ectopic pronephros, see Fig.1). At 1 ng enpp4 mRNA dose ectopic 3G8 (7%) and 4A6 (5%) staining were obtained only in rare cases (n = 94, Fig. 6d) compared to the optimized dose of enpp4 mRNA used in Fig. 1. Injection of any tested lipidic receptor mRNA alone does not induce any ectopic kidney formation with normal 3G8 and 4A6 staining in the majority of the embryos (n = 52, Fig. 6c, Fig. S6b, d, f). Only co-expression of s1pr5 and enpp4 mRNAs resulted significantly in higher ectopic 3G8 staining compared to enpp4 mRNA alone (38%, n = 89, p < 0.001, Fig. 6a, b, Supplementary Fig. 6a, c, e and Supplementary Table 5). Furthermore, the size and frequency of the ectopic pronephroi were higher than with injections of 2 ng of enpp4 alone (see Fig. 1b–e). These results indicate that, among the tested receptors, only S1pr5 enhanced Enpp4 function to generate ectopic pronephros.
Injected embryos were examined by 3G8/4A6 antibody staining following a, b double targeted injection of 2 ng s1pr5.l mRNA and 1 ng of enpp4 mRNA, c single targeted injection of s1pr5.l, d enpp4 mRNA or e, f embryos injected with 15 ng of s1pr5.L MO. g, h Double targeted injection of 7.5 ng of s1pr5.L MO and 5 ng of enpp4 MO1. i, j Single targeted injection of s1pr5.L MO (i) or enpp4 MO1 (j). k, l Double targeted injection of 15 ng of s1pr5.L MO and 2 ng of enpp4 mRNA (k) and 15 ng of cMO and 2 ng of enpp4 mRNA (l). An asterisk denotes the control uninjected side. An arrowhead indicates ectopic 3G8 staining (see also Supplementary Table 5 and Supplementary Figs. 6 and 7).
The functions of Enpp4 are mediated by the lipidic S1pr5 receptor
To further confirm that S1pr5 is involved in pronephros development we performed loss of function experiments. Two s1pr5 genes are identified in X.laevis genome and s1pr5.L corresponds to our published sequence32. Despite distinct spatial expression in the adult frog, the two s1pr5 homeologs display a very similar expression profile during X.laevis embryogenesis and are both expressed in the pronephric tissues (Supplementary Fig. 7a, b). We therefore performed loss of function analyses with an anti-sense MO against Xenopus s1pr5.L and s1pr5.S (see Supplementary Figs. 6g, h and 7c, d for MOs efficiency and specificity evaluation and Supplementary Table 5 for raw data and statistical analyses). Embryos injected with 15 ng of s1pr5.L MO or s1pr5.S MO displayed significant reduced 3G8 and 4A6 staining (n = 43 and 66, respectively, Fig. 6e, f, Supplementary Fig. 7e and Supplementary Table 5) suggesting that S1pr5 receptors are required for normal pronephros formation in Xenopus.
To examine potential synergistic effects, we co-injected 7.5 ng of s1pr5.L MO with 5 ng of enpp4 MO (half of the dose used previously for single injections) and compared their phenotype to those obtained following co-injection of s1pr5 MO or enpp4 MO with control MO (Fig. 6g–j). As expected, embryos co-injected with s1pr5.L and enpp4 MOs generated the strongest phenotype and smallest pronephros, with strong reduction of 3G8 (74%) and 4A6 (81%) staining domains (n = 42, Fig. 6g, h). Enpp4 MO alone also caused strongly reduced 3G8 (65%) and 4A6 (60%) staining as previously shown (n = 40, Fig. 6j), while the s1pr5.L MO alone reduced pronephric size in both 3G8 (22%) and 4A6 (29%) domains less frequently (n = 51, Fig. 6l). Although there are no significant differences in pronephric phenotype between enpp4 MO1 + s1pr5.L MOs and enpp4 MO1 injected embryos, we concluded that co-injection of s1pr5.L and enpp4 MOs showed additive effects on the inhibition of Xenopus pronephros development based on the size of the scored pronephroi.
To further analyse the link between Enpp4 and S1pr5, we carried out injection of 2 ng of enpp4 mRNA together with 15 ng of s1pr5.L MO (Fig. 6k–l) or control MO. As expected, injection of s1pr5.L MO lowered the percentage of embryos displaying ectopic 3G8 and 4A6 staining caused by enpp4 mRNA injection (17.5% and 0%, respectively, n = 40, Fig. 6k; compared to 48% and 10%, n = 50, Fig. 6l). Furthermore, the size and number per embryos of these ectopic pronephroi was lower than with injection of 2 ng of enpp4 and 15 ng of cMO. Moreover, the percentage of embryos injected with enpp4 mRNA and s1pr5.L MO displaying a reduced 3G8 and 4A6 expression domain remains high (42.5% and 30%, respectively, n = 40, Fig. 6k), most certainly due to the loss of function of S1pr5.
These results indicate that the ectopic pronephric tissues induced by Enpp4 overexpression are due to the activation of the S1pr5 receptor.
Enpp4 specifically interacts with the lysophospholipid phosphatidylserine
To assess if Enpp4 hydrolyses lipids and generates a ligand, which could bind to the S1pr5 receptor, phospholipid binding was tested by a protein-lipid overlay assay using commercial pre-spotted lipid membranes. Out of the 26 bioactive lipids tested, only phosphatidylserine (PS) is specifically bound by Xenopus Enpp4 (Fig. 7a, b and Supplementary Fig. 8). Moreover, this interaction is abolished when the putative catalytic site is mutated (Fig. 7c). We then tried to determine the enzymatic activity of Xenopus Enpp4. However, we could not detect any lipid derivatives, e.g. DAG, PA or LysoPS, which could be generated from the hydrolysis of PS in the membrane proteins fractions from overexpressing Enpp4 CHO cells. Taken together, these data show Enpp4 specifically interacts with PS but does not have PLA, PLC or PLD activity towards PS.
a Membrane Lipid stripTM was incubated with membrane protein extracts from Enpp4 overexpressing CHO cells and the bound Enpp4 protein detected with anti-XlEnpp4 serum. b, c Nitrocellulose membranes were spotted with increasing amount of PA or PS and incubated with membrane protein extracts from CHO cells transfected with enpp4-pcDNA3.1, with enpp4T72S-pcDNA3.1 or empty plasmid (control) and the bound proteins detected with anti-XlEnpp4 serum. LPA lysophosphatidic acid, LPC lysophosphocholine, PA phosphatidic acid, PC phospatidylcholine, PE phosphatidylethanolamine, PS phosphatidylserine, PtIns phosphatidylinositol, S1P shingosine-1-phosphate (see also Supplementary Fig. 8).
Discussion
This paper reports newly identified and unexpected roles of the conserved ectonucleotidase Enpp4 during vertebrate kidney development. Moreover, our findings provide a novel molecular mechanistic understanding for pronephric development and emphasizes the importance of the lipidic pathways in kidney formation (Fig. 8).
a During normal pronephric development, in the extracellular space, Enpp4 binds to phosphatidylserine close to, or in its catalytic site, which can then either interact with the S1pr5 or produce a novel ligand X, able to bind to this receptor. The activation of S1pr5 leads to the upregulation of lhx1/pax8 pronephric markers in the kidney field either by acting upstream of RA signalling pathway or by acting directly via the ERK or calcium pathways. At later stages, RA is required for tubules morphogenesis and Notch and Wnt pathway are involved in the patterning of the pronephric tubules. The mechanism by which S1pr5 activation directs the expression domains of these genes remains to be confirmed. b Enpp4 and S1pr5 overexpression lead to expanded and ectopic expression domains for both the Notch and RA pathway genes and wnt4. These changes in patterning gene expression domains induce the formation of enlarged pronephric segments and ectopic pronephric tubules.
We previously showed that enpp4 is expressed in pronephric tubules16, but our present data demonstrate that low but significant levels of enpp4 can be detected at the time of proximal tubule specification33. Weak enpp4 expression was previously detected by RT-PCR but not by ISH, in gastrula embryo16. This discrepancy is attributable to the lower sensitivity of ISH compared to RT-PCR for the detection of gene expression patterns.
Our work demonstrates that Enpp4 regulates the expression level of two of the transcription factors involved in pronephric anlagen formation, lhx1 and pax823 but not irx124. However, induction of lhx1 ectopic expression is delayed compared to pax8 one. Such a distinct expression regulation of these two pronephric genes by several signalling pathways has already been described34,35. As Lhx1 is necessary for the early patterning of the entire kidney and subsequently growth and elongation in the development of the pronephric tubules36,37, the reduction of lhx1 expression can explain the formation of the small pronephros in enpp4 morphants. Pax8 is necessary for the earliest steps of pronephric development and for pronephric precursors cell proliferation and can induce the formation of ectopic pronephric tubules23,38. Therefore, the ectopic expression of pax8 in Enpp4 overexpressing embryos can explain the formation of ectopic pronephroi. We also demonstrate that Enpp4 is sufficient to generate kidney, but only from lateral mesoderm and not in ectopic non-lateral positions. This suggests that the lateral mesoderm must contain either the receptor or the substrate necessary for Enpp4 function. Furthermore, the induction of enpp4 expression in activin treated animal caps confirms the importance of mesoderm tissues for Enpp4 pronephric functions (see Supplementary Fig. 4).
The ectopic kidneys formed from enpp4 overexpression consist of tubular structures and are patterned along their proximal/distal axis. Moreover, in some rare cases, there is a complete mini-duplication of the entire pronephros. This surprising phenotype could be explained by the upregulation of the patterning of signalling pathway members. RA signalling is required during gastrulation for pronephric specification. Increased levels of RA signalling by Enpp4 overexpression could lead to pax8 expression activation and then to the formation of ectopic pronephroi23,25. RA signalling is also required post-gastrulation for tubules morphogenesis and its downregulation in enpp4 morphants could explain tubules formation defects25. Subsequently, RA increase could pattern these ectopic tubules, as it has been shown during zebrafish pronephric nephron segmentation patterning39,40. However, in Xenopus pronephros, RA signalling increases expression level of distal tubules markers41. Moreover, RA signalling also regulates the expression of members of the Notch pathway42, which can subsequently activate wnt4 expression28, which then functions to pattern the proximal pronephros. Our data demonstrate that Rnfg protein is involved in mediating Enpp4 signalling, probably by its ability to modify Notch-ligand interactions43. Therefore, we speculate that Enpp4 acts upstream or in parallel to RA signalling and upstream of Notch and Wnt pathways (see Fig. 8). As enpp4 expression was unchanged in animal caps treated with RA compared to control caps, this supports the hypothesis that Enpp4 acts upstream of the RA pathway.
A key question is how the misexpression of Enpp4, an ectonucleotidase, can alter gene expression. Phosphatidylserine translocation across the cell membrane is a well-known indicator of apoptosis but is also involved in physiological and developmental processes44,45. Therefore, Enpp4 could bind to PS in the extracellular space during pronephrogenesis. Enpp4 enzymatic activity is essential for ectopic kidney formation, suggesting that renal alterations are due to an excess or shortage of Enpp4 generated products in the extracellular space inducing cell responses via the activation of the S1pr5 receptor. However, our data strongly suggest that the Enpp4 kidney phenotype is not linked to the bioactive lipids LPA or S1P. The fact that the observed kidney phenotype might be due to a non-catalytic effect of Enpp4 might be puzzling and unexpected, especially since Enpp6 has been suggested to play major renal physiological role through its enzymatic functions46. However, specific functions of other Enpps, such as Enpp1, Enpp2 and Enpp5, have been shown to be independent of their enzymatic activity47,48,49,50. It is therefore possible that Enpp4 does not hydrolyse PS but its interaction with PS is necessary for the activation of Enpp4 and subsequently of S1pr5. PS binding and conformational change mechanisms have been demonstrated for protein kinase C activation in mammalian kidney cells, supporting this hypothesis51. Although we were unable to detect any of the predicted products of PS hydrolysis, we cannot rule out that we failed to characterize Enpp4 enzymatic activity and that Enpp4 will generate a bioactive lipid, other than S1P, which is able to bind to S1p5r, the most divergent member of the S1pr family32. Although signalling through S1PR5 has been poorly studied, the activation of the S1P5 receptor has been linked to an intracellular calcium increase and inactivation of the ERK pathway, both pathways regulating pronephric field formation via RA signalling35,52,53,54,55.
We show that mouse Enpp4 can fulfil Xenopus Enpp4 functions during pronephrogenesis, suggesting mammalian kidney formation may be regulated by a similar mechanism demonstrated in this work. ENPP4 is highly expressed in human metanephros and kidney tumours and its expression increases in deceased donor kidney biopsies with delayed graft function after kidney transplantation (data from human protein atlas)56. Interestingly, ENPP4 is localized close to RUNX2 gene, whose mutations cause cleidocranial dysplasia (CCD)57,58. Furthermore, a child with CCD and crossed renal ectopia has been reported, and given our data, we can speculate that the ectopic kidney is attributable to ENPP4 locus alterations59.
We propose a potentially novel model of action of the lipidic pathway in kidney physiology, implicating either bioactive lipids distinct from LPA and S1P molecules or a novel non-catalytic interaction. The fact that a S1P receptor might be activated other than by S1P binding may explain the controversy regarding the beneficial actions of FTY720 in renal pathologies60. Moreover, our study raises potentially fascinating possibilities regarding regenerative therapies for renal diseases. As therapies for chronic renal failure are still lacking, the identification of a novel pathway enabling the generation of ectopic kidneys may provide useful insights to therapeutics that enhance human renal regeneration.
Methods
Ethics statement
The work was carried out under a UK Home Office-approved animal procedures project license and approved by the University of Warwick Biological Ethics Committee.
Enpp4 cloning and site-directed mutagenesis
The Xenopus enpp4 cDNA (Accession number: BC 079717) was cloned into pcDNA3.1. Mouse Enpp4 cDNA (Accession number: BC027749) was cloned into pCS2+ and pcDNA3.1. Site-directed mutagenesis of Xenopus enpp4 was performed using a PCR-based approach. For each mutant, 2 successive rounds of PCR were carried out using the Pfx polymerase (Invitrogen) following the manufacturer’s protocol and using the primers listed below. The first round of PCR, performed using the enpp4-pRNA3 plasmid as template, allowed the amplification of two fragments of the enpp4 coding region, one upstream and containing the desired mutation (underscored in the primer sequence) and the other downstream and containing the mutation, respectively. For this, one amplification was performed using the upstream primer carrying out the mutation and the primer ORF downstream containing the stop codon and the other amplification using the downstream primer carrying out the mutation and the primer ORF upstream containing the ATG codon. The two PCR products were then mixed and a third PCR was carried out using this mixture as template using the upstream and downstream ORF primers, carrying out the BamHI and EcoRI restriction sites, respectively (in italic in the primer sequence). The final PCR product was digested by BamHI and EcoRI and inserted into the pCS2+ vector. The presence of the correct mutation was confirmed by sequencing for each mutant. The mutant enpp4 cDNA was then extracted from the pCS2+ and cloned into the pcDNA3.1. All constructs were verified by sequencing.
mRNA synthesis and morpholino oligonucleotides
Capped mRNAs were synthesized using mMESSAGE mMACHINE Kits (Ambion) from linearized plasmids. Plasmids used were Xenopus enpp4-pRNA3 (clone BC079717); mouse Enpp4-pCS2+; Xenopus mutant enpp4-pCS2+; Xenopus s1p5r.L-pCS2+ (clone DC111014); Xenopus s1p1r-pCMV-Sport6 (clone BC074356); Xenopus p2y10-pCMV-Sport6 (clone BC084356) and Xenopus rfng-pCMV-Sport628. enpp4 MO1 (5′-atgaaaacccttccaaacatcttga-3′), enpp4 MO2 (5′-gaaatgtcacacacgcagctcctat-3′), enpp6 MO (5′-aacgtgctgtacttagccatgccac-3′), s1pr5.L MO (5′-catggtttcgtcaatcctttatttc-3′), s1pr5.S MO (5′-catggttcagtcaatgctttatctc-3′), rfng MO28 and standard control MO (cMO) were designed and supplied by GeneTools, LLC.
Embryo culture, dissection, microinjections and lineage staining
Xenopus embryos were staged according to Faber and Nieuwkoop61. Kidney and pronephric anlagen dissections were performed in Barth X33. Each individual sample was injected into the lateral marginal zone of a ventral-vegetal blastomere (V2) at the 8-cell stage to target the pronephros. Pilot experiments were carried out to determine the enpp4 and lipidic receptors mRNAs and MOs quantities to inject, based on their abilities to alter kidney development without affecting the overall morphology of the embryos. The rfng mRNA and MO dose was used as previously published28. The LacZ (250 pg) mRNA was used as a lineage tracer. LacZ mRNA was injected alone or in combination with standard MO as controls. Injected embryos were cultured to various developmental stages, fixed in MEMFA and stained for β-galactosidase activity (Red-Gal or X-Gal staining) to identify correctly targeted embryos. Only embryos that had normal pronephros formation on the uninjected side and correctly targeted β-galactosidase staining on the injected side were scored.
Analysis of molecular marker expression in embryos
Whole-mount immunohistochemistry was performed using 3G8 and 4A6 monoclonal antibodies as previously described20. Whole-mount in situ hybridization was carried out as previously described62. Anti-sense digoxigenin (DIG)-labelled RNA probes were synthesized from linearized template plasmids24,28,63. Either BM purple (Roche Applied Science) or NBT/BCIP (Roche Applied Science) or Fast Red/Napthol AS/MX (Sigma) was used for the colour reaction. After bleaching, embryos were photographed with a magnification of ×10 for whole stage 41 embryos, ×20 for whole gastrula, neurula and early organogenesis stages embryos and ×32 for pronephric region.
Acrylamide embedding, cryostat sectioning and Hoechst staining
X. laevis embryos were embedded sectioned at 18 µm thickness and nuclear Hoechst staining performed64.
RT-PCR
RT-PCR reactions were carried out on whole or dissected X. laevis embryos as described previously using the housekeeping gene odc as loading control. Quality of pronephric tissues dissections was assessed by amplification of the kidney markers lhx1 and pax8 and of the muscle marker myf5. Amplification conditions and primers sequences for the enpp, lpar, s1pr, lhx1 and pax8 genes have been previously published16,32,65. Myf5 was amplified using the forward primer, 5′-actactacagtctcccaggacaga-3′ and the reverse primer, 5′-agagtctggaatagggagggagca-3′, with the annealing temperature of 60 °C and 29 cycles. Each sample was analyzed in two independent embryo batches.
Cell culturing and transient transfection
Chinese hamster ovary (CHO) cells were cultured in HAMs F-12 (Gibco BRL) containing 10% foetal serum, NaHCO3 at 1.176 g/l, 2 mM of glutamine, 5 U/ml penicillin and 37.8 U/ml streptomycin during the 48 h prior to transfection. Cells were then transfected for 24 h using the reagent Turbofect (Fermentas) with 1 μg of the eukaryotic constructs. The transfection medium was then removed and replaced with culture medium. Approximately 48 h post transfection, cells were fixed or harvested for analyses. As a control, CHO cells were transfected with empty vector pcDNA3.1.
Anti-Enpp4 antibody production
The anti-Xl Enpp4 polyclonal antibody was raised in rabbits by direct intramuscular injection of the Xenopus laevis wild-type enpp4-pcDNA3.1 plasmid followed by electroporation (Aldevron, LLC, USA). Rabbits were immunized three times, at day 0, day 28 and 56 and terminal bleed performed at day 70.
Immunocytochemistry
Immunocytochemistry was carried out on fixed unpermeabilized cells with polyclonal antibodies anti-XlEnpp4 used at 1/200 or anti-mEnpp4 (CR65; see ref. 31) used at 1/400 and anti-rabbit IgG FITC (Sigma) at 1/80. The staining was recorded using a Nikon Optiphot/ Diginet camera system. Photographs were taken at a magnification of ×40.
Electrophoresis and western blot
Native membrane proteins were extracted from transfected cells using the ProteoExtract® Native Membrane Protein Extraction Kit (Calbiochem). Proteins from whole cells and from the membrane and soluble fractions were separated on a 12% SDS-PAGE protein gel. Part of the gel was stained with Coomassie blue and processed for mass spectrometry analysis (Pôle Protéomique, Plateforme Génomique Fonctionnelle de Bordeaux, Université de Bordeaux) and the other was transferred onto a PVDF membrane (Bio-Rad) overnight at 4 °C. To limit non-specific binding, the anti-XlEnpp4 antibody was pre-absorbed on untransfected CHO cells. The membrane was incubated overnight at 4 °C with 1:200 dilution of Enpp4 antiserum, washed and incubated in goat anti-rabbit IgG peroxidase secondary antibody (Sigma, dilution 1/2000) for 30 min at 20 °C. After several washes, immunoreactivity was detected by chemiluminescence (Western lighting Chemiluminescence Reagent Plus, Perkin Elmer).
Lipid binding assay
Hydrophobic membrane pre-spotted with bioactive lipids (ShingoStripsTM S-6000 and Membrane Lipids StripsTM S-6002; Echelon Biosciences) were blocked 1 h at 20 °C with 1% BSA in Tris-buffered saline 0.05% Tween-20 (TBST). All subsequent washes were performed in TBST. Blots were overlaid with proteins extracts from membrane fractions of CHO cells transfected with enpp4-pcDNA3.1 or empty vector (dilution 1/30) in blocking buffer overnight at 4 °C. Membranes were washed and incubated with pre-absorbed Enpp4 serum (dilution 1/200) for 6 h at room temperature. After several washes, the membranes were incubated with goat anti-rabbit IgG peroxidase secondary antibody for 30 min at room temperature, washed and developed using enhanced chemiluminescence. To confirm the observed binding, nitrocellulose Hybond-C extra (GE Healthcare) membranes were spotted with 0 to 200 µM of PA (Sigma P-9511) or PS (Sigma P-6641) diluted into a mix of MeOH/CHCl3/H2O (2/1/0.8, v/v). Dried membranes were then treated as described above.
Statistics and reproducibility
All experiments were repeated several times, on different batches of embryos, and pronephric phenotypes were determined in a commonly used way, blind-coded, by comparing the injected and uninjected sides28. The percentages of the embryos displaying the discussed phenotypes are given in the text in bracket along with the total number of analysed embryos. All raw data and statistical analyses are presented in the Supplementary Tables (SI). Each histological analysis was numbered (see Supplementary Tables 1a, 2a, 3a and 5a) and statistically pairwise compared as indicated in the Supplementary Tables 1b, 2b, 3b and 5b. Chi-square statistical analysis could not be performed for all comparison of experiments since the assumption was not always fulfilled. Hence, 2×5 Fisher’s exact test was therefore used. Bonferroni multiple testing correction was then applied to all statistical analyses. All the statistical analyses were performed using the R statistical software Core Team R66.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors confirm that all the data supporting the findings of this study are available in this article and its Supplementary Information files. Unedited gels and western blot are also presented in Supplementary Figs. 8 and 9.
References
Desgrange, A. & Cereghini, S. Nephron patterning: lessons from Xenopus, zebrafish, and mouse studies. Cells 4, 483–499 (2015).
Jones, E. A. Xenopus: a prince among models for pronephric kidney development. J. Am. Soc. Nephrol. 16, 313–321 (2005).
Lienkamp, S. S. Using Xenopus to study genetic kidney diseases. Semin. Cell Dev. Biol. 51, 117–124 (2016).
Wessely, O. & Tran, U. Xenopus pronephros development—past, present, and future. Pediatr. Nephrol. 26, 1545–1551 (2011).
Krneta-Stankic, V., DeLay, B. D. & Miller, R. K. Xenopus: leaping forward in kidney organogenesis. Pediatr. Nephrol. 32, 547–555 (2017).
Solini, A., Usuelli, V. & Fiorina, P. The dark side of extracellular ATP in kidney diseases. J. Am. Soc. Nephrol. 26, 1007–1016 (2015).
Park, F. & Miller, D. D. Role of lysophosphatidic acid and its receptors in the kidney. Physiol. Genomics 49, 659–666 (2017).
Huwiler, A. & Pfeilschifter, J. Sphingolipid signaling in renal fibrosis. Matrix Biol. 68-69, 230–247 (2018).
Burnstock, G., Evans, L. C. & Bailey, M. A. Purinergic signalling in the kidney in health and disease. Purinergic Signal 10, 71–101 (2014).
Dwyer, K. M., Kishore, B. K. & Robson, S. C. Conversion of extracellular ATP into adenosine: a master switch in renal health and disease. Nat. Rev. Nephrol. 16, 509–524 (2020).
Kirby, R. J. et al. Dynamic regulation of sphingosine-1-phosphate homeostasis during development of mouse metanephric kidney. Am. J. Physiol. Ren. Physiol. 296, F634–F641 (2009).
Kihara, Y., Maceyka, M., Spiegel, S. & Chun, J. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br. J. Pharm. 171, 3575–3594 (2014).
Burnstock, G. Purine and purinergic receptors. Brain Neurosci. Adv. 2, 2398212818817494 (2018).
Zimmermann, H., Zebisch, M. & Strater, N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal 8, 437–502 (2012).
Stefan, C., Jansen, S. & Bollen, M. NPP-type ectophosphodiesterases: unity in diversity. Trends Biochem. Sci. 30, 542–550 (2005).
Massé, K., Bhamra, S., Allsop, G., Dale, N. & Jones, E. A. Ectophosphodiesterase/nucleotide phosphohydrolase (Enpp) nucleotidases: cloning, conservation and developmental restriction. Int. J. Dev. Biol. 54, 181–193 (2010).
Albright, R. A. et al. NPP4 is a procoagulant enzyme on the surface of vascular endothelium. Blood 120, 4432–4440 (2012).
Albright, R. A. et al. Molecular basis of purinergic signal metabolism by ectonucleotide pyrophosphatase/phosphodiesterases 4 and 1 and implications in stroke. J. Biol. Chem. 289, 3294–3306 (2014).
Yan, D. et al. Homology modeling and docking studies of ENPP4: a BCG activated tumoricidal macrophage protein. Lipids Health Dis. 15, 19 (2016).
Vize, P. D., Jones, E. A. & Pfister, R. Development of the Xenopus pronephric system. Dev. Biol. 171, 531–540 (1995).
Raciti, D. et al. Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 9, R84 (2008).
Reggiani, L., Raciti, D., Airik, R., Kispert, A. & Brandli, A. W. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 21, 2358–2370 (2007).
Carroll, T. J. & Vize, P. D. Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev. Biol. 214, 46–59 (1999).
Alarcon, P., Rodriguez-Seguel, E., Fernandez-Gonzalez, A., Rubio, R. & Gomez-Skarmeta, J. L. A dual requirement for Iroquois genes during Xenopus kidney development. Development 135, 3197–3207 (2008).
Cartry, J. et al. Retinoic acid signalling is required for specification of pronephric cell fate. Dev. Biol. 299, 35–51 (2006).
Lyons, J. P. et al. Requirement of Wnt/beta-catenin signaling in pronephric kidney development. Mech. Dev. 126, 142–159 (2009).
McLaughlin, K. A., Rones, M. S. & Mercola, M. Notch regulates cell fate in the developing pronephros. Dev. Biol. 227, 567–580 (2000).
Naylor, R. W. & Jones, E. A. Notch activates Wnt-4 signalling to control medio-lateral patterning of the pronephros. Development 136, 3585–3595 (2009).
Saulnier, D. M., Ghanbari, H. & Brandli, A. W. Essential function of Wnt-4 for tubulogenesis in the Xenopus pronephric kidney. Dev. Biol. 248, 13–28 (2002).
Taelman, V., Van Campenhout, C., Solter, M., Pieler, T. & Bellefroid, E. J. The Notch-effector HRT1 gene plays a role in glomerular development and patterning of the Xenopus pronephros anlagen. Development 133, 2961–2971 (2006).
Moller, S. et al. Monitoring the expression of purinoceptors and nucleotide-metabolizing ecto-enzymes with antibodies directed against proteins in native conformation. Purinergic Signal 3, 359–366 (2007).
Massé, K., Kyuno, J., Bhamra, S. & Jones, E. A. The lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) receptor gene families: cloning and comparative expression analysis in Xenopus laevis. Int. J. Dev. Biol. 54, 1361–1374 (2010).
Brennan, H. C., Nijjar, S. & Jones, E. A. The specification of the pronephric tubules and duct in Xenopus laevis. Mech. Dev. 75, 127–137 (1998).
Drews, C., Senkel, S. & Ryffel, G. U. The nephrogenic potential of the transcription factors osr1, osr2, hnf1b, lhx1 and pax8 assessed in Xenopus animal caps. BMC Dev. Biol. 11, 5 (2011).
Futel, M. et al. TRPP2-dependent Ca2+ signaling in dorso-lateral mesoderm is required for kidney field establishment in Xenopus. J. Cell Sci. 128, 888–899 (2015).
Chan, T. C., Takahashi, S. & Asashima, M. A role for Xlim-1 in pronephros development in Xenopus laevis. Dev. Biol. 228, 256–269 (2000).
Cirio, M. C. et al. Lhx1 is required for specification of the renal progenitor cell field. PLoS ONE 6, e18858 (2011).
Buisson, I., Le Bouffant, R., Futel, M., Riou, J. F. & Umbhauer, M. Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development. Dev. Biol. 397, 175–190 (2015).
Naylor, R. W. et al. BMP and retinoic acid regulate anterior-posterior patterning of the non-axial mesoderm across the dorsal-ventral axis. Nat. Commun. 7, 12197 (2016).
Wingert, R. A. et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 3, 1922–1938 (2007).
Van Campenhout, C. et al. Evi1 is specifically expressed in the distal tubule and duct of the Xenopus pronephros and plays a role in its formation. Dev. Biol. 294, 203–219 (2006).
Arima, K. et al. Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Dev. Dyn. 232, 414–431 (2005).
Haines, N. & Irvine, K. D. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4, 786–797 (2003).
Leventis, P. A. & Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 (2010).
van den Eijnde, S. M. et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631–3642 (2001).
Sakagami, H. et al. Biochemical and molecular characterization of a novel choline-specific glycerophosphodiester phosphodiesterase belonging to the nucleotide pyrophosphatase/phosphodiesterase family. J. Biol. Chem. 280, 23084–23093 (2005).
Belfiore, A. et al. Overexpression of membrane glycoprotein PC-1 in MDA-MB231 breast cancer cells is associated with inhibition of insulin receptor tyrosine kinase activity. Mol. Endocrinol. 10, 1318–1326 (1996).
Dennis, J., Nogaroli, L. & Fuss, B. Phosphodiesterase-Ialpha/autotaxin (PD-Ialpha/ATX): a multifunctional protein involved in central nervous system development and disease. J. Neurosci. Res. 82, 737–742 (2005).
Grupe, A., Alleman, J., Goldfine, I. D., Sadick, M. & Stewart, T. A. Inhibition of insulin receptor phosphorylation by PC-1 is not mediated by the hydrolysis of adenosine triphosphate or the generation of adenosine. J. Biol. Chem. 270, 22085–22088 (1995).
Ohe, Y. et al. Characterization of nucleotide pyrophosphatase-5 as an oligomannosidic glycoprotein in rat brain. Biochem. Biophys. Res. Commun. 308, 719–725 (2003).
Bell, R. M., Hannun, Y. A. & Loomis, C. R. Mechanism of regulation of protein kinase C by lipid second messengers. Symp. Fundam. Cancer Res. 39, 145–156 (1986).
Im, D. S. et al. Characterization of a novel sphingosine 1-phosphate receptor, Edg-8. J. Biol. Chem. 275, 14281–14286 (2000).
Le Bouffant, R. et al. Retinoic acid-dependent control of MAP kinase phosphatase-3 is necessary for early kidney development in Xenopus. Biol. Cell 104, 516–532 (2012).
Leclerc, C., Webb, S. E., Miller, A. L. & Moreau, M. An increase in intracellular Ca2+ is involved in pronephric tubule differentiation in the amphibian Xenopus laevis. Dev. Biol. 321, 357–367 (2008).
Sanchez, T. & Hla, T. Structural and functional characteristics of S1P receptors. J. Cell Biochem. 92, 913–922 (2004).
Mas, V. R. et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation 85, 626–635 (2008).
Cohen, M. M. Jr. Perspectives on RUNX genes: an update. Am. J. Med. Genet. A 149A, 2629–2646 (2009).
Glusman, G., Kaur, A., Hood, L. & Rowen, L. An enigmatic fourth runt domain gene in the fugu genome: ancestral gene loss versus accelerated evolution. BMC Evol. Biol. 4, 43 (2004).
Suresh, S. S. A family with cleidocranial dysplasia and crossed ectopic kidney in one child. Acta Orthop. Belg. 75, 521–527 (2009).
Jo, S. K., Bajwa, A., Awad, A. S., Lynch, K. R. & Okusa, M. D. Sphingosine-1-phosphate receptors: biology and therapeutic potential in kidney disease. Kidney Int. 73, 1220–1230 (2008).
Faber, J. & Nieuwkoop, P. D. Normal Table of Xenopus laevis (Garland Publishing Inc, 1994).
Harland, R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 (1991).
Kyuno, J., Massé, K. & Jones, E. A. A functional screen for genes involved in Xenopus pronephros development. Mech. Dev. 125, 571–586 (2008).
Seville, R. A., Nijjar, S., Barnett, M. W., Massé, K. & Jones, E. A. Annexin IV (Xanx-4) has a functional role in the formation of pronephric tubules. Development 129, 1693–1704 (2002).
Haldin, C. E. et al. The lmx1b gene is pivotal in glomus development in Xenopus laevis. Dev. Biol. 322, 74–85 (2008).
Core Team R. R. in R Foundation for Statistical Computing (http://www.R-project.org/) (2013).
Acknowledgements
We are grateful to Junichi Kyuno for his contribution to experimental design and data analysis. We thank E. Pera for the raldh1a2, ralhd10 and cyp26a1 in situ probes, A. Tocco for technical help and P. Jarrett for maintenance of frogs. We also thank J-W. Dupuy (Pôle Protéomique Plateforme Génomique Fonctionnelle Bordeaux, Université de Bordeaux) for MS analysis, and B. Arveiler for helpful comments regarding human pathologies. The work was supported by Wellcome trust grant 082071 as well as CNRS, University of Bordeaux and ANR.
Author information
Authors and Affiliations
Contributions
K.M. and E.A.J. designed research; K.M., E.A.J., S.B. and L.M.-P. performed research; K.M., E.A.J., C.P., E.B.-G. and L.M.-P. analysed data; K.M. and E.A.J. wrote the paper with contributions from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Edwina McGlinn and Luke R. Grinham. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Massé, K., Bhamra, S., Paroissin, C. et al. The enpp4 ectonucleotidase regulates kidney patterning signalling networks in Xenopus embryos. Commun Biol 4, 1158 (2021). https://doi.org/10.1038/s42003-021-02688-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02688-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.