Abstract
Staphylococcus pseudintermedius is a commensal bacterium and a major opportunistic pathogen of dogs. The emergence of methicillin-resistant S. pseudintermedius (MRSP) is also becoming a serious concern. We carried out a population genomics study of 130 clinical S. pseudintermedius isolates from dogs and cats in the New England region of the United States. Results revealed the co-circulation of phylogenetically diverse lineages that have access to a large pool of accessory genes. Many MRSP and multidrug-resistant clones have emerged through multiple independent, horizontal acquisition of resistance determinants and frequent genetic exchange that disseminate DNA to the broader population. When compared to a Texas population, we found evidence of clonal expansion of MRSP lineages that have disseminated over large distances. These findings provide unprecedented insight into the diversification of a common cutaneous colonizer of man’s oldest companion animal and the widespread circulation of multiple high-risk resistant clones.
Similar content being viewed by others
Introduction
Staphylococcus pseudintermedius is a commensal bacterium of the skin and mucous membrane and is frequently found in companion animals. Carriage of S. pseudintermedius often reaches >80% in some populations of healthy dogs1. It is also an opportunistic pathogen responsible for severe and necrotizing infections, and is frequently isolated in the skin, ears, bones, and post-surgical abscesses1. S. pseudintermedius has been found in other animals such as cats and horses, although they are not considered its natural hosts2. The bacterium shares several features with Staphylococcus aureus, the most important staphylococcal species in humans, including the capacity to express a range of virulence factors such as coagulase and other proteolytic enzymes, as well as a variety of toxins such as haemolysins, exfoliative toxins, enterotoxins, and leucotoxins1,3. The emergence of methicillin-resistant S. pseudintermedius (MRSP) is becoming a serious concern in veterinary medicine4,5,6 and highlights the need for accurate long-term surveillance. Furthermore, there have been recent reports of S. pseudintermedius being isolated in human carriage and infections, mainly associated with contacts with dogs (e.g., pet owners, veterinary staff)7,8. Although traditionally S. pseudintermedius has not been considered a risk for humans, recent reports have suggested that it is emerging as a zoonotic diagnosis and it may have been previously misidentified as S. aureus in human infections9,10.
Microbial population genomics involves sequencing the genomes of hundreds or even thousands of closely related strains within and between environments11. It has been instrumental in revolutionizing the epidemiology, surveillance and control strategies of infectious diseases that threaten human health, and has been widely used to investigate common bacterial species such as S. aureus, Klebsiella pneumoniae and Streptococcus pneumoniae12. Remarkable levels of genomic variation have been widely reported in bacterial pathogen populations, which have critical implications in understanding the origins of highly virulent and resistant lineages, successful colonization, transmission, and instances of host switching13,14,15. However, the application of large-scale whole-genome sequencing of bacterial species that naturally inhabit animal hosts remains limited. Even more problematic is when strains with multiple resistant phenotypes become prevalent in the population, but the lack of precise diagnostic and surveillance techniques makes them invisible to clinical and epidemiological studies. This gap in our knowledge ultimately limits our understanding of the underlying genetic characteristics of bacterial lineages that cause infections in animals and impacts the likelihood of precisely identifying new and emerging clones, including potential zoonoses.
The few genomic studies of S. pseudintermedius have offered a glimpse of its diversity, epidemiological characteristics and antimicrobial resistance (AMR) profile. A study of 12 genomes revealed that the multidrug-resistant and MRSP phenotypes in sequence types (ST) 71 and 68 in Europe evolved through a stepwise accumulation of mec-carrying chromosomal cassette (SCCmec), transposon Tn5405 and single-nucleotide polymorphisms (SNPs) conferring fluoroquinolone resistance4. In the Netherlands, S. pseudintermedius is dominated by STs 45, 71, and 258, and genome sequencing of 50 isolates showed different resistance profiles among members of each clone5. These results suggest that the AMR phenotype is acquired and maintained through both vertical inheritance and horizontal gene transfer (HGT)5. In New Zealand, ST 71-dominated MRSP population has undergone infrequent ancestral acquisition events of the mobile chromosomal cassette SCCmec, which was followed by its subsequent widespread dispersal6. However, genomic studies aimed at elucidating the structure and dynamics of S. pseudintermedius populations remain scarce. Such information will be critical to addressing the question of whether the multidrug-resistant and MRSP phenotypes of S. pseudintermedius are restricted to one or few clones, the factors that drive their evolution, and how the species will respond to different selective pressures. It will also help us clarify the breadth of potentially useful adaptive variants that the species possesses and the underlying mechanisms that allow it to successfully switch between hosts. In this study, we carried out a population genomics analysis of 130 clinical S. pseudintermedius isolates from dogs and cats across five states in the New England region of the United States sampled from 2017 to 2018. Results reveal the diversification and widespread circulation of multiple high-risk resistant clones.
Results
Diverse lineages are co-circulating in New England
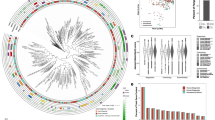
We obtained a total of 162 S. pseudintermedius isolates from routine diagnostic tests of clinical specimens submitted to the New Hampshire Veterinary Diagnostic Laboratory (NHVDL) from October 2017 to October 2018. Of the 162 S. pseudintermedius isolates, we retrieved high-quality draft genome sequences for 130 isolates, composed of 126 genomes from dogs and four from cats (Supplementary Fig. 1 and Supplementary Data 1). More than half of the isolates came from New Hampshire (n = 84), whereas the remainder were from neighboring states in New England (Massachusetts, Connecticut, Maine, and Vermont). Initial in vitro screening of methicillin and other beta-lactam antibiotic resistance was performed using disc diffusion testing. Results revealed the presence of 36 MRSP isolates and 94 methicillin-susceptible S. pseudintermedius (MSSP) isolates throughout the entire study period. We found MRSP isolates in almost every month of sampling, with as many as six out of 12 isolates being MRSP in a single month (Fig. 1a).
a Monthly sampling of MRSP and MSSP isolates based on results of disk diffusion test. b The midpoint-rooted maximum likelihood phylogenetic tree was calculated using sequence variation in 1906 core genes. Scale bar represents nucleotide substitutions per site. For visual clarity, not all months of sampling are shown on the color legend of month heat map; however, all months are shown on the middle strip. c Frequency distribution of pairwise genome-wide ANI values. d Gene presence–absence matrix showing the distribution of AMR determinants present in each genome (blue blocks–present; white–absent). AMR determinants refer to both horizontally acquired genes and allelic variants of a gene. e Distribution of the number of AMR determinants per genome. f Gene presence–absence matrix showing the distribution of virulence-associated genes present in each genome (red blocks–present; white–absent). Each column in the two matrices correspond to a genome on the phylogeny. NH New Hampshire, MA Massachusetts, CT Connecticut, ME Maine, VT Vermont.
De novo genome assembly generated sequences of sizes ranging from 2.44 to 2.93 Mb (mean = 2.61 Mb) and the number of predicted genes ranged from 2260 to 2850 (mean = 2449) (Supplementary Data 1). We used Roary16 to estimate the pan-genome of the entire New England S. pseudintermedius population. Roary identified a total of 8207 orthologous gene families in the pan-genome, which can be classified into core genes (n = 1906 genes), soft core genes (n = 108 genes), shell genes (n = 741 genes), and cloud genes (n = 5452) (Supplementary Fig. 2 and Supplementary Data 2). The combined core and soft core genes comprised 24.5% of the pan-genome, whereas the combined shell and cloud genes (which together make up the accessory genome) comprised 75.5% of the pan-genome. It is notable that many accessory genes were unique to a single strain (2797 genes, representing 34.1% of the pan-genome). The large accessory genome in our results is not uncommon and has been reported in different bacterial pathogens, including S. aureus17,18 and Staphylococcus haemolyticus19. However, we recognize that pan-genome sizes and precise characterization of core and accessory genes are greatly influenced by the phylogenetic distance, sequence quality of genomes, the number of genomes being compared, assembly and annotation methods, and choice of threshold values to define orthologous genes. The maximum likelihood phylogenetic tree based on the sequence alignment of the 1906 core genes reveal many deep branching lineages that have relatively little structure relative to sampling location (state) or month of sampling (Fig. 1b). The four isolates from cats are intermingled among the dog isolates throughout the tree. To further ensure that all isolates are from the same species and determine the degree of overall genomic relatedness among the S. pseudintermedius isolates, we calculated the pairwise average nucleotide identity (ANI) of all orthologous genes shared between any two genomes20. Genome-wide ANI values for every possible pair of S. pseudintermedius genomes range from 98.8–100% (mean = 99.3%) (Fig. 1c, Supplementary Fig. 3 and Supplementary Data 3).
We considered bioinformatic evidence for the presence of different AMR determinants in our samples. We used ARIBA to detect the presence of horizontally acquired AMR genes and resistance alleles owing to chromosomal mutations21. The distribution of AMR determinants varied substantially among the genomes and we did not find evidence for phylogenetic or geographical clustering of any one of the AMR genes (Fig. 1d and Supplementary Data 4). Two mechanisms confer penicillin resistance in Staphylococcus: the production of beta-lactamase encoded by blaZ, which inactivates penicillin by hydrolysis of its beta-lactam ring, and the penicillin-binding protein PBP2a encoded by mecA22. We detected blaZ in 111 isolates representing 85.4% of the population, and the mecA gene in 32 genomes representing 24.6% of the population. The genes tetK and tetM, which confer resistance to broad-spectrum tetracyclines, were detected in three and 59 genomes, representing 2.3% and 45.4% of the population, respectively. We also detected resistance determinants for other antimicrobial classes. Many genomes carry genes that encode resistance against aminoglycosides (aac(6)-aph(2), aph(3')-III, ant(6')-Ia, sat4a; n = 38, 38, and 39 genomes, respectively), chloramphenicols (cat; n = 12 genomes), daptomycin (rpoB allelelic variant; 17 genomes), diaminopyrimidines (dfrG, dfrK; n =31 and 6 genomes, respectively), fusidic acid (fusA allelelic variant; 99 genomes), lincosamides/macrolides/streptogramin B (lnuG, ermB; n = 3 and 34 genomes, respectively), and streptothricin (sat4; n = 38 genomes). Only four isolates out of the 130 genomes do not contain any known AMR determinant, whether it is a resistance allele or a horizontally acquired gene. Although none of the isolates from the four cats are MRSP, all four genomes harbor DNA that encodes resistance against other antibiotic classes. Overall, we found that a total of 126 S. pseudintermedius genomes carry at least one AMR determinant, and a remarkable 41 genomes contain ≥5 AMR determinants per genome (Fig. 1e).
Mobile genetic elements (MGEs) play an important role in the evolution and diversity of Staphylococcus and facilitate the dissemination of AMR genes within and between species23. Here, we explored the presence and types of the chromosomal cassette SCCmec, which can facilitate the mobilization and distribution of mecA and other AMR genes in Staphylococcus24,25. SCCmec elements are highly variable in terms of their structural organization and gene content, but are classified mainly based on the ccr and mec gene complexes, the key elements of the cassette responsible for integration and excision of SCCmec and the beta-lactam resistance phenotype, respectively26. A total of 13 SCCmec types and numerous subtypes have been identified in S. aureus to date26. Of the 32 S. pseudintermedius genomes that carry the SCCmec, we identified three known types (Type III (also known as Type II–III in MRSP27), n = 2 genomes; Type IV, n = 10 genomes; Type V, n = 15 genomes) and five genomes with unknown type (Supplementary Fig. 4 and Supplementary Data 1). These results were similar to those identified in veterinary MRSP in Australia27. Their locations in the phylogenetic tree show that the MRSP phenotype has been derived from at least 13 independent acquisitions of different SCCmec elements. In S. aureus, Type III is found predominantly in hospital-associated methicillin-resistant S. aureus (MRSA) and carry resistance genes for cadmium, tetracycline, mercury, erythromycin, and spectinomycin, in addition to mecA28. Hence, a single transfer event of Type III can turn a susceptible strain into a multidrug-resistant strain. Types IV and V are often found in community-associated MRSA and carry only the mecA gene28. In S. pseudintermedius, SCCmec III tended to be healthcare-associated MRSP lineages, whereas isolates with SCCmec V tended to be community-associated MRSP lineages29. The five untypeable SCCmec highlight the need to continue efforts to discover and characterize undiscovered variants of SCCmec elements, especially in non-aureus species.
We identified several virulence genes commonly found in S. aureus in the New England S. pseudintermedius population (Fig. 1f and Supplementary Data 5). The two-component pore-forming leukocidin genes lukF and lukS30 were present in all genomes. The synergistic action of these genes produces a toxin that damages membranes of host defense cells and erythrocytes3,31. Genes associated with other virulence factors were also detected from the genomic sequences, although present in lower frequencies. These include enterotoxins encoded by sec (n = 15 genomes), selK (n = 11 genomes), selI (n = 15 genomes), and selq (n = 12 genomes). Staphylococcal enterotoxins encoded by sec and sel cause superantigenic and emetic activities in the bacterium31.
Overall, we found high levels of phylogenetic and genomic diversity in S. pseudintermedius isolates co-circulating in New England only within a single year of sampling. The population consists of several methicillin-resistant, multidrug-resistant, and virulent clones present in dogs and cats.
Comparison of two S. pseudintermedius populations
To place our data set in a country-wide context, we compared the genome sequences of New England S. pseudintermedius to a previously published genomic dataset from Texas32 (Fig. 2a, Supplementary Fig. 3 and Supplementary Data 1). New England, located in the northeast corner of the United States, is ~2770 km from Texas, which is in the southern part of the country (Fig. 2b). We chose this data set because its sampling strategy was most similar to our study, i.e., only isolates from clinical specimens from dogs collected by the Texas Veterinary Medical Teaching Hospital and a mix of MRSP and MSSP32. Of the 160 Texas genomes sequenced in that study, we did not include genomes of isolates from healthy dogs and those with low-quality sequences. To minimize the confounding effect of the host animal, we also excluded the four isolates from cats in the New England data set. Overall, we compared 126 New England and 107 Texas genomes, all from disease cases in dogs.
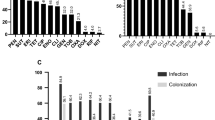
All isolates in this tree were sampled from clinical specimens from dogs submitted to the veterinary laboratory in each region. a Midpoint-rooted maximum likelihood tree built using concatenated sequence alignments of 1776 core genes present in both New England and Texas populations. Scale bar represents nucleotide substitutions per site. Upside down triangles indicate the presence of mecA in genome sequences. Blue bar plots represent number of horizontally acquired AMR genes per genome. b Map of the United States showing the New England and Texas regions. c Distribution of STs per region. Comparison between the New England and Texas populations in terms of the total number of AMR genes per genome d, percentage of each population that carry specific AMR genes e, percentage of the population that carry at least one AMR gene for each major class of antibiotic f, and the total number of genomes that are resistant to zero, one or multiple classes of antibiotics g. Red horizontal lines in d and g represent median values. e, f Only statistically significant differences are shown for visual clarity. g Counts of genomes are based on the presence of at least one AMR gene that represent each major class of antibiotic.
We next re-ran Roary on each data set. The composition of the pan-genome varied between the two populations. The New England population has a total of 8122 genes (1910 core, 107 soft core, 765 shell, 5340 cloud genes), whereas the Texas population has a total of 6845 genes (1836 core, 133 soft core, 1020 shell, 3856 cloud genes) (Supplementary Data 2). The combined New England and Texas data set has a total of 9817 genes (1776 core, 166 soft core, 905 shell, 6970 cloud genes) (Supplementary Fig. 5 and Supplementary Data 2). We used the concatenated sequences of 1776 core genes that are common to both New England and Texas populations, generating a 1.60 Mb sequence alignment, to build a maximum likelihood phylogeny (Fig. 2a). We observed frequent intermingling of isolates from each population across the tree, suggesting widespread dissemination across a large geographical distance.
We carried out a second AMR analysis to compare horizontally acquired AMR genes in the two populations using ABRicate (Supplementary Data 6). In this analysis, we did not use ARIBA because ARIBA requires the raw reads as input, which were not available for the Texas genomes. Based on in silico detection of the mecA gene, ABRicate identified 44 MRSP and 63 MSSP in the Texas population (Fig. 2a). In both New England and Texas populations, the MRSP lineages are mostly clonal, indicated by the extremely short branches at the tip of each clonal branch. The most common MRSP STs that had representatives from both regions were STs 45, 64, 150, and 155. There is also evidence for clonal expansion of some MRSP lineages within each region. For example, STs 68, 71, and 84 were found only in Texas, whereas ST 181 was found only in New England. Moreover, in both populations, the MSSP lineages are genetically diverse and are found in particularly long branches. There were also numerous STs that were either rare (here, defined as present in only two genomes) or unique (present only in a single genome) (Fig. 2c). STs that are intermingled with another may reflect recent divergence or recombination of the MLST genes.
Many genomes of the eight most common MRSP STs and their close relatives also carry the highest number of horizontally acquired AMR genes per genome, ranging from 2 to 11 AMR genes (median = 9 AMR genes per genome) (Supplementary Data 6). We did not find significant differences between the two populations in terms of the total number of AMR genes per genome (p value = 0.14, Welch's t test) (Fig. 2d). We further queried the genomes to compare the distribution of specific AMR genes between New England and Texas (Fig. 2e). Both had comparable proportions of genomes that carry many of these AMR genes, such as aac-aph2, blaZ, and tetM. However, we found significant differences between the two populations in those genomes that carry the gene ermB (p = 0.0233, z score test) and mecA (p = 0.0054, z score test). For both genes, the Texas population exhibited higher number of genomes as a proportion of their respective population size. In terms of the major classes of antibiotics, both populations have high numbers of genomes that carry genes conferring resistance against beta-lactams, but they differ in the proportions of genomes that carry resistance genes for macrolides (p = 0.0233, z score test), streptogramin B (p = 0.0233, z score test) and diaminopyrimidines (p = 0.0475, z score test) (Fig. 2f). We also found that a total of 45 genomes representing 35.7% of the New England population and 48 genomes representing 44.9% of the Texas population carry resistance genes to ≥3 antibiotic classes (Fig. 2g). We did not detect any horizontally acquired AMR genes in 15/126 (11.9%) and 10/107 (9.35%) of the New England and Texas genomes, respectively.
Within-species diversity stems from allelic differences in core genes as well as gene content variation in the accessory genomes among strains14, which may partly stem from differential response to local selective pressures through frequent DNA gain and loss13,14. We therefore sought to determine to what extent the genome structures of S. pseudintermedius in New England and Texas have diverged. First, using the 1.60 Mb core genome alignment, we calculated the total number of SNPs between each pair of genomes from each population. We did not find significant differences in the number of SNPs in the core genome (p value = 0.46, Welchʼs t test) (Fig. 3a), but found significant differences in the number of accessory genes per genome between the two populations (p value < 0.00016, Welch's t test) (Fig. 3b). To gain insight on how the accessory genome has diverged in relation to the core genome14, we used PopPUNK, which employs pairwise nucleotide k-mer comparisons with distinguish shared sequence and gene content33. We found similar patterns in the distribution of pairwise genomic distances between New England and Texas, with larger genetic distances concentrated away from the origin (Fig. 3c). This pattern is indicative of the presence of multiple genetically distinct clusters that are diverging in both core sequences and accessory gene content. These results expand the core genome-based phylogenetic analysis described above (Fig. 2a). Overall, these data show that each of the two geographically distinct populations of S. pseudintermedius in the United States is composed of numerous genetically distinct lineages that can be distinguished in both their core and accessory genome divergence patterns.
a Pairwise core SNPs. b Number of accessory genes per genome. Red horizontal lines represent median values. c Contour plots of pairwise distances between genomes in terms of their core genome divergence (measured by SNP density across the core genome) and the difference in their accessory genomes (measured by the Jaccard distance based on the variation in the gene content of their sequences).
Heterogeneity in the frequency of recombination
Many bacterial species are known to receive DNA fragments from other taxa and integrate them into their genomes through recombination34. The process of recombination has a fundamental role in the evolution of many bacterial pathogens and has been implicated in the emergence of new phenotypic traits such as AMR, ecological adaptation, and host colonization34. Recombination is known to contribute to the evolution of Staphylococcus, as in the case of S. aureus and Staphylococcus epidermidis, which have been reported to frequently recombine within and between species35. Here, we sought to determine the frequency and characteristics of recombination in S. pseudintermedius because this process may likely contribute to its genomic variation and diversification.
We used different methods to elucidate the impact of recombination on the population structure and evolution of S. pseudintermedius. First, under the null hypothesis of no recombination, we calculated the pairwise homoplasy index (PHI)36 and detected evidence for significant recombination in the core genome of the combined New England and Texas data set (p = 0.0, permutation test). Using the sequence alignment of the 1776 core genes shared by the New England and Texas genomes, we used the SplitsTree network37 to visualize the reticulated, star-like evolutionary history of the combined S. pseudintermedius populations (Fig. 4a). We next sought to identify frequently recombining genes in each population by running fastGEAR38 on individual sequence alignments of core and shared accessory genes. The program fastGEAR identifies both recent (i.e., recombination events affecting a few strains) and ancestral (i.e., recombination events affecting entire lineages) recombinations (Fig. 4b, Supplementary Data 7). In the New England population, we found that a total of 961 genes have had a history of recombination. A total of 922 and 199 genes were involved in recent recombination and ancestral recombination, respectively. In contrast, we identified a total of 869 genes that have experienced recombination in the Texas population, with 799 and 184 genes that have experienced recent and ancestral recombination, respectively. Although many of the frequently recombined genes have hypothetical or unknown functions, we found that copB, which has a role as a copper-translocating P-type ATPase39, is frequently recombined in both populations. In S. aureus, this gene is part of the operon that promotes copper hypertolerance and enhanced resistance to phagocytic killing39. Metal resistance genes, including copB, are carried by some SCCmec variants in both human and animal MRSA and other species of Staphylococcus40. Hence, copB is likely co-translocated and/or co-selected with AMR genes in the SCCmec. Another gene that has experienced frequent recombination is fnbA, which encodes a fibronectin-binding protein and thus promotes colonization to different anatomical sites of the eukaryotic host41. The gene bca, which encodes the surface-associated C protein alpha antigen42, is also frequently recombined in both populations. In S. aureus, sequence variation in fnbA has been previously reported to contribute to differences in antigenicity between strains41,43.
a A phylogenetic SplitsTree network of the core genome. Scale bar represents nucleotide substitutions per site. b Genes that have undergone recent and ancestral recombination. Horizontal axis shows the estimated number of ancestral recombinations, and vertical axis shows the estimated number of recent recombinations. Names of some of the most frequently recombined genes with known functions are shown. List of all recombined genes are shown in Supplementary Data 7. c–g Evolutionary and recombination parameters calculated by mcorr.: c recombination coverage; d diversity; f mean fragment size of a recombination event; ϕ recombinational divergence; θ mutational divergence; γ/μ relative rate of recombination to mutation (equivalent to ratio of ϕ/θ). Red vertical line in each plot represents the inferred value of each parameter calculated with 1000 bootstrapped replicates.
We also wanted to compare different genome-wide evolutionary and recombination parameters between New England and Texas populations. We used the core genome alignment of each data set as an input to mcorr44 (Fig. 4c–g and Supplementary Data 8). The recombination coverage (c), which indicates the fraction of the genome whose diversity was derived from recombination events since the last common ancestor of the population, was estimated to be 0.274 (standard deviation [sd] = 0.014) and 0.210 (sd = 0.013) in New England and Texas, respectively. Recombination coverage ranges from 0 (clonal evolution) to 1 (complete recombination)44. These values indicate that 27.4% and 21.0% of sites in any one genome from New England and Texas, respectively, originated from recombination events. The diversity (d) is the probability that a pair of genomes will differ at any locus and is estimated from the diversity generated from both recombination and accumulation of mutations of the clonal lineage44. This parameter was estimated to be 3.88 × 10−3 (sd = 8.80 × 10−5) and 3.46 × 10−3 (sd = 8.73 × 10−5) in New England and Texas, respectively. The mean fragment size (f̅) of a recombination event was estimated to be 426.14 (sd = 60.22) bp and 387.09 (sd = 40.55) bp in New England and Texas, respectively. Recombinational divergence (ϕ) was estimated to be 0.060 (sd = 0.009) and 0.055 (sd = 0.006) in New England and Texas, respectively. The mutational divergence (θ), which refers to the mean number of mutations per locus since divergence of a pair of homologous sites in the pool, was estimated to be 0.014 (sd = 6.39 × 10−4) and 0.016 (sd = 9.48 × 10−4) in New England and Texas, respectively. Lastly, the ratio ϕ/θ (or γ/μ), which gives the relative rate of recombination to mutation, was estimated to be 4.40 (sd = 0.683) and 3.52 (sd = 0.413) in New England and Texas, respectively. For all parameters, we found significant differences between the two populations (p value <0.001 for all six parameters; Welch's t test). For comparison, S. aureus has lower values for recombination divergence (0.042) and ϕ/θ (1.0), and higher values for recombination coverage (0.36), diversity (0.015), mean fragment size (550 bp), and mutational divergence (0.042)44. Overall, we found that heterogeneity in rates and characteristics of recombination has contributed to shaping the genomic structure and diversity of the New England and Texas populations of S. pseudintermedius.
Discussion
We present a comparative population genomics study of S. pseudintermedius in the United States. Results revealed the co-circulation of phylogenetically diverse lineages that have access to a large pool of accessory genes, including AMR genes. For S. pseudintermedius, the long-term clinical and public health significance of MRSP and multidrug-resistant lineages remains unclear. Suffice to say that companion animals can act as a reservoir of resistant strains that can be transmitted to humans or other animals, as has been reported for other Staphylococcus species that colonize multiple eukaryotic hosts45,46. S. pseudintermedius also appears to be undergoing a considerable amount of genetic exchange, which can lead to the emergence of high-risk clones and those with hitherto uncharacterized virulence capacity. The origins of recombined and horizontally acquired DNA remains unclear, but they were likely derived from other S. pseudintermedius strains, other Staphylococcus species that they co-inhabit their niche with, and even distantly related organisms. This has been demonstrated in a recent study that reported that many resistance genes have been frequently exchanged between different Staphylococcus species and between Staphylococcus and other non-Staphylococcus taxa47. The ability of S. pseudintermedius to take up and integrate large DNA segments highlights the pathogen's capacity for rapid evolution and adaptation to primary and secondary hosts, similar to what has been observed in S. aureus35,46. Frequent recombination may also contribute in part to the pathogenicity of S. pseudintermedius and its ability to colonize other host species. Recombination is not the only process that contributes to pathogen diversity. Extensive phylogenetic intermixing of strains spanning a large geographical distance suggests substantial translocation and/or exchange of strains. This may likely be a result of either widespread pet transport and distribution across the country or that there is a country-wide pool of circulating MSSP and MRSP lineages, with indication of local expansion and diversification of certain MRSP clones in each region.
A few limitations need to be acknowledged. The retrospective nature of the isolate collection we used and sampling bias owing to the inclusion of only clinical submissions mean that sampling may have missed some less-prevalent genotypes and counties in New England. We also did not have access to data regarding the antibiotic history, treatment received, or clinical outcomes of the dogs and cats from which isolates were obtained. Another limitation is our use of databases for querying AMR genes, virulence genes and SCCmec types. These databases were developed primarily for S. aureus, of which thousands of genomes have already been sequenced (for example, see ref. 46). Less is known of other Staphylococcus species, however, and this gap in current knowledge may have important consequences. For example, newly discovered genetic variants (as in the case of the five unknown SCCmec types) may be present in other species but remain invisible from current in silico detection methods and databases, which make them difficult to track over the long term. This also means that clinical decisions and treatment options made when treating diseases caused by non-aureus species are based on assumptions from studies of S. aureus. Estimates of evolutionary and recombination parameters show considerable differences not only between the two S. pseudintermedius populations but also between S. pseudintermedius and S. aureus, which can impact population dynamics and the emergence of favorable phenotypes. Hence, more extensive and large-scale genome sequencing of non-aureus species will greatly augment this gap in our knowledge of the diversity, pathogenicity, and resistance mechanisms of other Staphylococcus species that can potentially develop into formidable high-risk pathogens. Notwithstanding these limitations, we obtained sufficient representation of the diversity of S. pseudintermedius and the widespread distribution of MRSP and multidrug-resistant clones even within a few years of sampling, which was the main focus of this study.
The results presented here open multiple avenues for future research. First, the integration of genomic epidemiology in veterinary medicine will advance our ability to identify targets for strain differentiation, surveillance, and diagnostics. It is imperative that long-term monitoring of the eight major STs with the highest number of horizontally acquired AMR genes (STs 45, 64, 68, 71, 84, 150, 151, and 181) should be carried out, as they appear to be expanding in New England and Texas and likely in other parts of the country. Future work using functional assays is also needed to precisely characterize the virulence factors of S. pseudintermedius, instead of simply relying on S. aureus studies. Our results on the most frequently recombining genes (e.g., copB) provides an important starting point to understand the genetic basis of S. pseudintermedius pathogenicity. Second, local and global surveillance of resistant phenotypes is particularly critical to detect and limit the emergence of new resistant clones with the capacity to spread and switch between animals and humans. Future work should include determining the prevalence of MRSP and multidrug-resistant strains across a range of domestic and wildlife animals, and specific groups of people who are in constant proximity to animals (e.g., veterinary personnel, pet owners, breeders, pet shop owners, pet adoption workers, farmers). Close contact between animals and humans has facilitated multiple host-switching events in S. aureus48,49,50 and it is not unreasonable to assume that it can also occur in S. pseudintermedius. Investigation into the extent of host switching (i.e., animal-to-human and between dogs and other animal species) and the adaptive changes associated with radical changes in host ecology will reveal key host–pathogen interactions that could be targeted for novel clinical interventions and therapies. Third, the precise origins of MRSP (whether from human-associated methicillin-resistant S. aureus, methicillin-susceptible animal-associated S. pseudintermedius or other staphylococcal species) remains unclear and genomic sequencing approaches will certainly advance our understanding of how and when it emerged in companion animals. We also encourage the application of population genomics as an integral component of the One Health initiative, which emphasizes that the health of humans, animals, and the environment are inextricably linked51. Taken together, our findings provide a genomic framework that will provide a critical foundation and practical support for future studies, investigating the population structure and dynamics, drug resistance and diversification of S. pseudintermedius as well as the risks to pets, pet owners, and veterinary personnel. It will facilitate more deeply informed genomic tracking and surveillance for the emergence and convergence of virulence and AMR in certain genotypes of S. pseudintermedius and in different geographical regions. In summary, we underscore the value of elucidating the population genomic structure and evolution of this increasingly important pathogen to advance the health of man’s favorite companion and oldest domestic animal.
Methods
Bacterial sample collection
The S. pseudintermedius isolates were acquired as culture swabs from routine clinical specimen submissions to NHVDL, Durham, New Hampshire, USA, from October 2017 to October 2018. The clinical specimens were received from veterinary practices in the New England region that includes the states of Connecticut, New Hampshire, Maine, Massachusetts, and Vermont. No live vertebrates were used in this study; therefore, the NHVDL is exempt from the IACUC approval process. All isolates were from disease cases. Pure isolates of S. pseudintermedius were cultured on commercially prepared tryptic soy agar with 10% sheep red blood cells. Initial species identification was carried out using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), which has been previously demonstrated to be a sensitive and specific species-level identification tool for S. pseudintermedius6,32. We used the Bruker Biotyper MALDI-TOF MS following manufacturer’s protocols. The addition of formic acid in the extended direct technique was used when necessary to ensure a reliable log(score), which refers to the level of similarity between an unknown tested specimen and a reference sample. Species assignments were made when log(score) values were ≥2.0 and identical species were included in the top two database matches when compared with the library of reference spectra available in the Bruker MBT Compass, RUO library 6903(V6) and 7311(V7) (Bruker Daltonics, Bremen, Germany). The most common sites of isolation included skin, ears, urine, and wounds. Metadata information of isolates are displayed in Supplementary Data 1. All isolates were stored in DMSO solution in −80 °C.
Methicillin susceptibility screening
Isolates were initially screened in vitro at the NHVDL with the Kirby Bauer disc diffusion technique using both the cefoxitin and oxacillin discs. Whereas cefoxitin predicts methicillin resistance in S. aureus, oxacillin is used as the official predictor of methicillin resistance for S. pseudintermedius following breakpoint guidelines of the most current Clinical and Laboratory Standards Institute52. For methicillin-resistant isolates, we further tested for the presence of the penicillin-binding protein PBP2 using a commercial latex agglutination test (MASTALEX MRSA Latex Kit, MAST, UK) following manufacturer's protocols. Verification for the presence of the mecA gene was done using whole-genome sequencing (described below).
DNA extraction and whole-genome sequencing
Cultures were grown in brain heart infusion broth at 37 °C for 24–48 h. DNA was extracted and purified from cultures using the Zymo Research Quick-DNA Fungal/Bacterial Miniprep Kit (Irvine, California) following manufacturer's protocols. DNA concentration was measured using a Qubit fluorometer (Invitrogen, Grand Island, NY). DNA libraries were prepared using the Nextera XT protocol (as per the manufacturer's instructions) with 1 ng of genomic DNA/isolate. Samples were sequenced as multiplexed libraries on the Illumina HiSeq platform operated per the manufacturer's instructions to produce paired end reads of 250 nucleotides in length. Sequencing was done at the UNH Hubbard Center for Genome Studies, Durham, NH, USA.
Genome assembly, annotation, and pan-genome analysis
Reads were assembled into contigs using the de novo assembler SPAdes v.3.13.153. The resulting contigs were annotated using Prokka, a stand-alone tool specifically developed for annotating bacterial genomes54. Genome assembly quality was assessed using Quast55. Sequencing failures and poor overall sequence qualities are known to occur with any sequencing platform. We used CheckM to assess the quality of our sequences and excluded genomes with <90% completeness and >5% contamination56. After filtering out the genomes with low coverage and of poor quality and exclusion of any assemblies with >200 contigs and an N50 <40,000 bp, we used a total of 130 genomes for downstream analysis, with the numbers of contigs ranging from 25 to 196 and N50 ranging from 50,676 to 351,605 bp (Supplementary Fig. 1 and Supplementary Data 1). We also confirmed the species identity using a BLASTN57 search of the NCBI non-redundant (nr) database using the annotated 16S rDNA sequences of each genome in our study. To determine the degree of genomic relatedness and hence clarify whether these genomes belong to the same species, we calculated the genome-wide ANI for all possible pairs of genomes using the program FastANI v.1.020. ANI is a robust similarity metric that has been widely used to resolve strain relatedness and determine whether any two strains belong to the same or different species20. We used the program Roary16 to characterize the pan-genome of the 130 New England S. pseudintermedius. Roary iteratively pre-clusters protein sequences using CD-HIT58, all-against-all BLASTP57 and Markov clustering59. Roary then sorts all orthologous gene families identified in the pan-genome into four categories: core (genes found in ≥99% strains), soft core (found in 95% ≤strains <99%), shell (found in 15% ≤strains <95%), and cloud (found in >15% of strains)16. We used the default parameters for all programs used.
Comparative population genomics of New England and Texas
We compared 126 New England genomes with 107 previously published genomes from Texas for a total of 233 genomes32. For both data sets, we included only those clinical isolates from dogs (disease cases). The four genomes from cats in the New England data set were excluded. A list of accession numbers and metadata for the Texas S. pseudintermedius genomes were included in Supplementary Data 1. To maintain consistency in gene annotations, we used Roary to characterize the pan-genome of this merged data set and re-annotated them using Prokka with default parameters54. The US map used in the design of Figs. 1 and 2 was obtained from https://simplemaps.com.
Phylogeny and population structure analyses
For each of the two data sets (i.e., the 130 New England genomes and the 233 combined New England+Texas genomes), each single-copy orthologous gene family obtained from the Roary analysis was aligned using MAFFT60. The alignments were concatenated to give a single core alignment, and a maximum likelihood phylogeny was then generated using the program Randomized Axelerated Maximum Likelihood (RAxML) v.8.2.1161 with a general time-reversible nucleotide substitution model62 and four gamma categories for rate heterogeneity. Genetic population structure analysis was performed using R-implemented hierarchical Bayesian analysis of population structure (RhierBAPS) with default parameters using nested clustering and with the core genome alignment as input63. All phylogenies were visualized using the Interactive Tree of Life64. To further elucidate the population structure of S. pseudintermedius based on the divergence of both shared sequence and gene content in a population, we used PopPUNK (Population Partitioning Using Nucleotide K-mers) with default parameters33. PopPUNK compares all possible pairs of genomes by calculating the proportion of shared k-mers of different lengths to determine core and accessory genome distances. It then generates a scatterplot of the two distances to reveal the predicted clustering of isolates33.
In silico molecular typing and detection of AMR genes
ST identification of isolates was confirmed using the program MLST (https://github.com/tseemann/mlst), which extracts the sequences of seven housekeeping genes (tuf, cpn60, pta, purA, fdh, ack, sar65) from the contig files and compares them with the S. pseudintermedius MLST database (https://pubmlst.org/spseudintermedius/). We screened all of the genomes for known accessory element resistance genes using two programs. For the New England data set, we used a direct read mapping approach implemented in ARIBA66 that can identify resistance determinants owing to allelic variants and horizontally acquired AMR genes. For the combined New England and Texas genomes, we used the contig-based search method ABRicate v.0.8.13 (https://github.com/tseemann/abricate). The resistance allele sequences used for comparison were retrieved from the CARD database67. A query gene is accepted as a true AMR gene if it reaches a threshold of 95% sequence identity and 95% coverage when compared to the database. We used the default parameters for each program.
Detection of virulence genes and SCCmec
We screened all New England genomes for known virulence genes using ARIBA to query the Virulence Factor Database (VFDB)68. Genomes carrying the mecA-carrying chromosomal cassette SCCmec were identified using SCCmecFinder v.1.226 with minimum thresholds of >60% for sequence coverage and >90% sequence identity. We used the default parameters for each program.
Estimating recombination rates
We used three approaches to detect recombination in S. pseudintermedius. First, we calculated the PHI to determine the statistical likelihood of recombination being present in our data set36. If recombination is absent, the genealogical correlation or similarity of neighboring sites remains unchanged to permutations of the sites as all sites have identical evolutionary history36. We then visualized potential recombination events using Splitstree v.4.14.6, which shows the presence of conflicting phylogenetic signals for different genes37. Second, we ran fastGEAR38 with default parameters to detect evidence for recombination in core genes and shared accessory genes. FastGEAR first infers the population structure of individual sequence alignments using a Hidden Markov Model to identify lineages, which are groups of strains that are genetically distinct in at least 50% of the alignment38. Within each lineage, recombinations are identified by determining sequence similarity between the target sequence and all other remaining lineages38. Third, we used mcorr with default parameters to calculate the correlation profile and different evolutionary parameters using the core gene alignment as input and with 1000 bootstrapped replicates44. Mcorr estimates six evolutionary parameters: θ – mutational divergence; ϕ – recombinational divergence; c – recombination coverage or proportion of sites in the genome whose diversity was derived from outside the sample through recombination; d – diversity; mean fragment size (f̅) of a recombination event; and θ/ϕ (or γ/μ) – relative rate of recombination to mutation44.
Statistics and reproducibility
We used Welch's t test and z score test to compare the different genomic components between the New England and Texas populations. Significance of the inferred PHI was obtained using a permutation test36. We used a diversity test implemented in fastGEAR to determine the significance of the inferred recombinations and uncover false positives38. We used Welch's t test to compare the two S. pseudintermedius populations for each of the six parameters calculated by mcorr44. A p value of <0.05 was considered statistically significant. We used the default parameters for all programs used unless otherwise stated and are then described. Exact sample sizes for each group were described. Source data used to plot Figs. 2d–g, 3a, b, 4b–h are archived in Supplementary Data 9.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All supporting data are included in this published article and its supplementary material, and are available from the corresponding author upon request. Genome sequence data of the New England samples have been deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA563147 with BioSample accession numbers listed in Supplementary Data 1. Allelic profiles of the 79 STs previously unidentified to our knowledge from the New England genomes were submitted to the S. pseudintermedius database in the MLST website (https://pubmlst.org/spseudintermedius/). Accession numbers of the Texas genomes32 are listed in Supplementary Data 1 and have been downloaded from the PATRIC database (https://patricbrc.org). Source data underlying plots shown in figures are provided in Supplementary Data 9.
Code availability
All tools and R packages used for the analysis are publicly available and fully described in the “Method” sections.
References
Bannoehr, J. & Guardabassi, L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol. 23, 253–266 (2012). e51-52.
Ruscher, C. et al. Prevalence of methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet. Microbiol. 136, 197–201 (2009).
Maali, Y. et al. Understanding the virulence of Staphylococcus pseudintermedius: a major role of pore-forming toxins. Front. Cell Infect. Microbiol 8, 221 (2018).
McCarthy, A. J. et al. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 70, 997–1007 (2015).
Wegener, A. et al. Comparative genomics of phenotypic antimicrobial resistances in methicillin-resistant Staphylococcus pseudintermedius of canine origin. Vet. Microbiol. 225, 125–131 (2018).
Nisa, S. et al. Combining MALDI-TOF and genomics in the study of methicillin resistant and multidrug resistant Staphylococcus pseudintermedius in New Zealand. Sci. Rep. 9, 1271 (2019).
Van Hoovels, L., Vankeerberghen, A., Boel, A., Van Vaerenbergh, K. & De Beenhouwer, H. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 44, 4609–4612 (2006).
Paul, N. C., Moodley, A., Ghibaudo, G. & Guardabassi, L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health 58, 533–539 (2011).
Börjesson, S., Gómez-Sanz, E., Ekström, K., Torres, C. & Grönlund, U. Staphylococcus pseudintermedius can be misdiagnosed as Staphylococcus aureus in humans with dog bite wounds. Eur. J. Clin. Microbiol. Infect. Dis. 34, 839–844 (2015).
Somayaji, R., Priyantha, Ma. R., Rubin, J. E. & Church, D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn. Microbiol. Infect. Dis. 85, 471–476 (2016).
Sheppard, S. K., Guttman, D. S. & Fitzgerald, J. R. Population genomics of bacterial host adaptation. Nat. Rev. Genet. 19, 549–565 (2018).
Tagini, F. & Greub, G. Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur. J. Clin. Microbiol. Infect. Dis. 36, 2007–2020 (2017).
Chewapreecha, C. et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309 (2014).
Croucher, N. J. et al. Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat. Commun. 5, 5471 (2014).
Andam, C. P. et al. Genomic epidemiology of penicillin-nonsusceptible pneumococci with nonvaccine serotypes causing invasive disease in the United States. J. Clin. Microbiol. 55, 1104–1115 (2017).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Bosi, E. et al. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl Acad. Sci. USA 113, E3801–3809 (2016).
Park, S.-C., Lee, K., Kim, Y. O., Won, S. & Chun, J. Large-scale genomics reveals the genetic characteristics of seven species and importance of phylogenetic distance for estimating pan-genome size. Front. Microbiol. 10, 834 (2019).
Pain, M., Hjerde, E., Klingenberg, C. & Cavanagh, J. P. Comparative genomic analysis of Staphylococcus haemolyticus reveals key to hospital adaptation and pathogenicity. Front. Microbiol. 10, 2096 (2019).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018).
Hunt, M. et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Micro. Genom. 3, e000131 (2017).
Llarrull, L. I., Fisher, J. F. & Mobashery, S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob. Agents Chemother. 53, 4051–4063 (2009).
Firth, N., Jensen, S. O., Kwong, S. M., Skurray, R. A. & Ramsay, J. P. Staphylococcal plasmids, transposable and integrative elements. Microbiol. Spectr. 6, GPP3-0030 (2018).
International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53, 4961–4967 (2009).
Rolo, J. et al. Evolutionary origin of the Staphylococcal Cassette Chromosome mec (SCCmec). Antimicrob. Agents Chemother. 61, pii: e02302-16 (2017).
Kaya, H. et al. SCCmecFinder, a web-based tool for typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3, e00612-17 (2018).
Worthing, K. A. et al. Characterization of Staphylococcal cassette chromosome mec elements from methicillin-resistant Staphylococcus pseudintermedius infections in Australian animals. mSphere 3, pii: e00491-18 (2018).
Lakhundi, S. & Zhang, K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, pii: e00020-18 (2018).
Kasai, T., Saegusa, S., Shirai, M., Murakami, M. & Kato, Y. New categories designated as healthcare-associated and community-associated methicillin-resistant Staphylococcus pseudintermedius in dogs. Microbiol. Immunol. 60, 540–551 (2016).
Abouelkhair, M. A., Bemis, D. A., Giannone, R. J., Frank, L. A. & Kania, S. A. Characterization of a leukocidin identified in Staphylococcus pseudintermedius. PLoS ONE 13, e0204450 (2018).
Pinchuk, I. V., Beswick, E. J. & Reyes, V. E. Staphylococcal enterotoxins. Toxins (Basel) 2, 2177–2197 (2010).
Little, S. V., Bryan, L. K., Hillhouse, A. E., Cohen, N. D. & Lawhon, S. D. Characterization of agr groups of Staphylococcus pseudintermedius isolates from dogs in Texas. mSphere 4, e00033-19 (2019).
Lees, J. A. et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 29, 304–316 (2019).
Vos, M. Why do bacteria engage in homologous recombination? Trends Microbiol. 17, 226–232 (2009).
Méric, G. et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol. 7, 1313–1328 (2015).
Bruen, T. C., Philippe, H. & Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681 (2006).
Huson, D. H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73 (1998).
Mostowy, R. et al. Efficient inference of recent and ancestral recombination within bacterial populations. Mol. Biol. Evol. 34, 1167–1182 (2017).
Zapotoczna, M. et al. Mobile-genetic-element-encoded hypertolerance to copper protects Staphylococcus aureus from killing by host phagocytes. MBio 9, e00550-18 (2018).
Argudín, M. A. & Butaye, P. Dissemination of metal resistance genes among animal methicillin-resistant coagulase-negative Staphylococci. Res. Vet. Sci. 105, 192–194 (2016).
Loughman, A. et al. Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 8, 74 (2008).
Michel, J. L. et al. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89, 10060–10064 (1992).
Murai, M., Moriyama, H., Hata, E., Takeuchi, F. & Amemura-Maekawa, J. Variation and association of fibronectin-binding protein genes fnbA and fnbB in Staphylococcus aureus Japanese isolates. Microbiol. Immunol. 60, 312–325 (2016).
Lin, M. & Kussell, E. Inferring bacterial recombination rates from large-scale sequencing datasets. Nat. Methods 16, 199–204 (2019).
Guardabassi, L., Schwarz, S. & Lloyd, D. H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54, 321–332 (2004).
Richardson, E. J. et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2, 1468–1478 (2018).
John, J., George, S., Nori, S. R. C. & Nelson-Sathi, S. Phylogenomic analysis reveals the evolutionary route of resistant genes in Staphylococcus aureus. Genome Biol. Evol. 11, 2917–2926 (2019).
Weinert, L. A. et al. Molecular dating of human-to-bovid host jumps by Staphylococcus aureus reveals an association with the spread of domestication. Biol. Lett. 8, 829–832 (2012).
Shepheard, M. A. et al. Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS ONE 8, e62369 (2013).
Spoor, L. E. et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio 4, e00356-13 (2013).
Destoumieux-Garzón, D. et al. The One Health concept: 10 years old and a long road ahead. Front. Vet. Sci. 5, 14 (2018).
CLSI Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. VET01. (Clinical and Laboratory Standards Institute, 2018).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Tavaré, S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. Lectures Math. Life Sci. 17, 57–86 (1986).
Tonkin-Hill, G., Lees, J. A., Bentley, S. D., Frost, S. D. W. & Corander, J. RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 3, 93 (2018).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Solyman, S. M. et al. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J. Clin. Microbiol. 51, 306–310 (2013).
Inouye, M. et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6, 90 (2014).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Liu, B., Zheng, D., Jin, Q., Chen, L. & Yang, J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692 (2019).
Acknowledgements
The authors thank the UNH Resource Computing Center where all bioinformatics analyses were performed. We thank Anthony Westbrook for providing technical and bioinformatics assistance. This research was funded by UNH COLSA start-up funds. We thank the staff of NHVDL and the UNH Hubbard Center for Genome Studies for laboratory and technical support.
Author information
Authors and Affiliations
Contributions
C.P.A. designed the work. J.T.S. performed all bioinformatics analyses. J.T.S., S.A., C.J.M., D.N., and R.G. performed all sampling, culturing, and DNA extractions. C.P.A. and J.T.S. wrote the manuscript. C.P.A. guided the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Smith, J.T., Amador, S., McGonagle, C.J. et al. Population genomics of Staphylococcus pseudintermedius in companion animals in the United States. Commun Biol 3, 282 (2020). https://doi.org/10.1038/s42003-020-1009-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-020-1009-y
This article is cited by
-
First report of Staphylococcus pseudintermedius ST71-SCCmec III and ST45-ΨSCCmec57395 from canine pyoderma in Argentina
BMC Research Notes (2023)
-
Whole genome sequencing and pan-genome analysis of Staphylococcus/Mammaliicoccus spp. isolated from diabetic foot ulcers and contralateral healthy skin of Algerian patients
BMC Microbiology (2023)
-
Diagnostic and commensal Staphylococcus pseudintermedius genomes reveal niche adaptation through parallel selection of defense mechanisms
Nature Communications (2023)
-
Investigation of antimicrobial susceptibility and genetic diversity among Staphylococcus pseudintermedius isolated from dogs in Rio de Janeiro
Scientific Reports (2023)
-
Evolution and dispersal of mitochondrial DNA haplogroup U5 in Northern Europe: insights from an unsupervised learning approach to phylogeography
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.