Abstract
The phloem transports photosynthetic assimilates and signalling molecules. It mainly consists of sieve elements (SEs), which act as “highways” for transport, and companion cells (CCs), which serve as “gates” to load/unload cargos. Though SEs and CCs function together, it remains unknown what determines the ratio of SE/CC in the phloem. Here we develop a new culture system for CC differentiation in Arabidopsis named VISUAL-CC, which almost mimics the process of the SE–CC complex formation. Comparative expression analysis in VISUAL-CC reveals that SE and CC differentiation tends to show negative correlation, while total phloem differentiation is unchanged. This varying SE/CC ratio is largely dependent on GSK3 kinase activity. Indeed, gsk3 hextuple mutants possess many more SEs and fewer CCs, whereas gsk3 gain-of-function mutants partially increase the CC number. Taken together, GSK3 activity appears to function as a cell-fate switch in the phloem, thereby balancing the SE/CC ratio.
Similar content being viewed by others
Introduction
Multicellular organisms possess a variety of functional cells with a proper ratio for their life maintenance. In plants, the phloem tissue is composed of two major cell types: sieve elements (SEs) as conductive tubes and phloem companion cells (CCs) as helper cells for phloem transport. Phloem CCs function to support neighboring SEs through connected plasmodesmata. Although they function together with each other to ensure phloem transport, it has long been a deep mystery how the ratio of SE/CC is strictly controlled in the phloem. Recent technical advances enabled to identify various regulators that control SE differentiation1. In contrast to SEs, understanding of the molecular mechanism underlying CC differentiation remains a long-standing challenge.
Vascular cell induction culture system using Arabidopsis leaves (VISUAL) is a culture system that can artificially mimic plant vascular development2. In the VISUAL system, mesophyll cells are reprogrammed into vascular stem cells, and then differentiated into xylem vessel elements or phloem SEs within a couple of days. VISUAL enables the molecular genetic studies of vascular development, leading to the in-depth understanding of the regulatory network, especially for phloem SE differentiation. Even in VISUAL, differentiation into phloem CCs rarely occurs2, which makes it difficult to study CC development in detail.
In this study, we develop a new culture system for inducing CC-like cell differentiation named VISUAL-CC by modifying the conventional VISUAL method. Based on comprehensive gene expression analysis in VISUAL-CC, here we reveal that GLYCOGEN SYNTHASE KINASE 3 (GSK3) activity plays an important role in determining the SE/CC ratio. In vivo genetic analyses confirm the importance of GSK3 probably as a cell-fate switch in phloem development.
Results
VISUAL-CC is a culture system for inducing CC-like cells
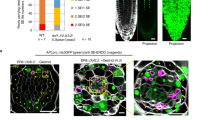
The conventional VISUAL system can induce ectopic xylem (XY) or phloem SEs via the stage of vascular stem cell (Fig. 1a). Toward the understating of CC differentiation, we modified the VISUAL based on a luciferase-based screen with SUCROSE-PROTON SYMPORTER 2 (SUC2) pro:ELUC, a specific CC marker3 (Fig. 1b). In this screen process, vascular stem cells were induced by the conventional VISUAL method in advance, and subsequently were exposed to a variety of culture media. After a series of screens with different media, we could induce pSUC2:ELUC activity (Fig. 1c) and ectopic expression of the pSUC2:YFPnls marker in cotyledons within 4 days using CC medium (Fig. 1d–f). Hereafter, we refer to this culture system as “VISUAL-CC”. To further investigate the spatial pattern, a dual phloem marker line expressing pSUC2:YFPnls and SIEVE-ELEMENT-OCCLUSION-RELATED 1 (SEOR1) pro:SEOR1-RFP5, a specific SE marker was established. Detailed observation of the dual phloem marker line by confocal microscopy after tissue-clearing treatment (ClearSee)4 revealed that CCs expressing pSUC2:YFPnls (green) are detected only in dividing cell clusters and are always limited to the cells adjacent to SEs expressing pSEOR1:SEOR1-RFP5 (magenta) (Fig. 1g). Thus, CC and SE markers appeared next to each other after several rounds of cell division (Fig. 1g). Observations using a field-emission scanning electron microscope (FE-SEM) or transmission electron microscopy (TEM) consistently visualized CC-like cells with dense cytoplasm adjacent to SEs with brighter cytoplasm (Fig. 1h). These cells showed minor vacuolation and developed the branched plasmodesmata typically seen in SE–CC complexes in vivo (Fig. 1h–j). In VISUAL, SMXL4 and SMXL5 are known as important regulators for early phloem SE development (Supplementary Fig. 1a)6,7. In VISUAL-CC, the double mutant smxl4 smxl5 significantly suppressed CC-like cell differentiation (Supplementary Fig. 1b), suggesting that SE and CC differentiation partially shares a common developmental process from vascular stem cells. Taken together, these results suggest that VISUAL-CC can mimic the SE–CC complex formation.
a The process of vascular cell differentiation in conventional VISUAL system. b Schematic of the screening system. LUC activity of pSUC2:ELUC was monitored every 20 min during culture under various conditions. c Time course of pSUC2:ELUC intensities during culture in conventional VISUAL and VISUAL-CC. The vertical axis indicates photon counts per second detected by the luminometer. The lower panel illustrates culture conditions and medium composition during VISUAL and VISUAL-CC culture; see Materials for a detailed protocol. d, e pSUC2:YFPnls expression before (d) and after 4 days of VISUAL-CC treatment (e) in cotyledons. f Simultaneous observation of pSUC2:YFPnls expression and xylem cells after VISUAL-CC treatment. Xylem cells were detected as UV autofluorescent signal (blue). g Expression patterns of pSEOR1:SEOR1-RFP (magenta) and pSUC2:YFPnls (green) in VISUAL-CC. Cell walls were stained with calcofluor white (blue). h FE-SEM image of ectopic CC-like cells (high electron density) and SEs (low electron density) induced by VISUAL-CC. i TEM image of branched plasmodesmata between an ectopic CC-like cell and a SE. j Schematic of branched plasmodesmata. Scale bars: 500 μm (d–g); 5 μm (h); 200 nm (i).
VISUAL-CC can induce known CC-related gene expression
To validate the promoter-based assay, we compared SUC2 mRNA accumulation with the promoter:LUC activity in the same sample (Fig. 2a, b). qRT-PCR analyses of VISUAL-CC samples and samples cultured in the conventional VISUAL medium (VISUAL, V) as negative controls revealed a strong correlation between promoter activity and mRNA level of SUC2 (Fig. 2b; r = 0.97, P < 0.005) (Fig. 2b). We used these data for classification of VISUAL-CC samples into strong LUC activity (CC-strong, S) and moderate LUC activity (CC-moderate, M), according to their SUC2 levels (Fig. 2a, b; Supplementary Fig. 2). Expression of SISTER OF ALTERED PHLOEM DEVELOPMENT (SAPL), another CC marker gene8, also showed a strong correlation with SUC2 expression (Fig. 2c; r = 0.91, P < 0.005). A microarray analysis using the same samples was performed to obtain a comprehensive gene expression profile (Supplementary Fig. 3a). As expected, genes previously characterized as CC-related, including SULFATE TRANSPORTER 2;1 (SULTR2;1)9, SODIUM POTASSIUM ROOT DEFECTIVE 1 (NaKR1)10, C-TERMINALLY ENCODED PEPTIDE RECEPTOR (CEPR1)/XYLEM INTERMIXED WITH PHLOEM 1 (XIP1)11,12, and MYB-RELATED PROTEIN 1 (MYR1)13, showed similar expression patterns to SUC2 and SAPL (Fig. 2d). Consistently, quantitative RT-PCR assay for these genes validated the microarray result (Supplementary Fig. 4). By utilizing the variation in expression observed between samples (S, M, and V), we identified 186 VISUAL-CC-inducible genes that satisfied the following patterns of expression levels: S > M > V and S/V > 4 (Fig. 2e and Supplementary Fig. 2). According to the previous dataset of root cell-type-specific transcriptome14, these genes were mainly expressed in root CCs or phloem pole pericycles (PPPs, Fig. 2f). PPPs are also known to participate in phloem unloading from SEs in roots via intervening plasmodesmata15 (Fig. 2f). Here we grouped 67 genes as VISUAL-CC-related (VC) genes based on CC-preferential expression (Supplementary Table 1). Transporter genes were overrepresented among these VC genes, reflecting the functional aspect of phloem transport (Supplementary Fig. 3b, c).
a Time course of pSUC2:ELUC signal intensities for each category: CC-strong (S), CC-moderate (M), and VISUAL (V) (n = 5). Error bars indicate SD. b Correlation between pSUC2:ELUC signals and relative SUC2 mRNA expression levels for each sample. c Correlation of expression levels between SUC2 and SAPL in S and M samples. d Expression levels of known CC-related genes in each sample. Relative expression levels were calculated when expression in S3 was set to 1. e Heat map of expression levels of VISUAL-CC-inducible genes in each sample. Expression data were normalized against the median value and are presented according to the color scale (log2) above the chart. f Cluster analysis of VISUAL-CC-inducible genes based on a previous root cell-type-specific transcriptome dataset14. A schematic of the primary phloem tissue patterning found in roots is shown on the left-hand side of the chart. g–i Heat map of expression levels of VC (67 genes, g), VS (218 genes, h), and VS (137 genes, i) in S and M samples. The lower panel indicates the mean value for each sample. j Heat map of expression levels of 113 bikinin-suppressed genes in S and M samples.
SE and CC differentiation shows a negative correlation
We previously identified 137 VISUAL-XY-related (VX) genes and 218 VISUAL-SE-related (VS) genes using VISUAL microarray data2. Then, expression of VX and VS genes was examined in the VISUAL-CC transcriptome dataset. Although there was no regular pattern of VX gene expression, expression of VS genes was very low in the S samples in contrast to that of VC genes (Fig. 2g–i). Correlation analysis among these gene sets revealed that expression levels of VC genes negatively correlate with those of VS genes (Supplementary Fig. 5b; r = −0.91, P < 0.05) but not with those of VX genes (Supplementary Fig. 5a; r = −0.28, P > 0.05). To further assess this tendency, we calculated the quantitative expression levels of vascular marker genes in individual samples. Although there was a strong correlation between expression of SAPL (CC) and SUC2 (CC) (Fig. 2c, r = 0.91, P < 0.005), no correlation was found between IRREGULAR XYLEM 3 (IRX3)16 (XY) and SUC2 (CC) expression (Fig. 3a, r = −0.27, P > 0.05). By contrast, expression of SEOR1 (SE) showed a significant negative correlation with that of SUC2 (CC) (Fig. 3b, r = −0.76, P < 0.05). All these results suggest that CC and SE differentiation tend to show negative correlation. Interestingly, expression levels of ALTERED PHLOEM DEVELOPMENT (APL) (SE + CC), which is expressed in both SEs and CCs17, was almost constant among all the samples, indicating that the total amount of differentiating phloem cells is almost unchanged (Fig. 3c and Supplementary Fig. 6a). Taken together, these results suggest that VISUAL-CC induces different ratios of CC-like cells and SEs without changing the total number of phloem cells, while VISUAL only produced SEs. This implies that a key determinant of SE or CC cell fate is present in VISUAL-CC cultures.
a Expression of SUC2 (CC) and IRX3 (XY) showing no correlation between levels in S and M samples. The Pearson correlation coefficient and P value are marked on the chart. b Negative correlation between expression of SUC2 (CC) and SEOR1 (SE). c Positive correlation between expression of SUC2 (CC) and APL (SE + CC). d Time course of pSUC2:ELUC signal intensities during VISUAL-CC with various bikinin concentrations. Averaged LUC values are shown (n = 6). e–i Expression of SUC2 (CC, e), SAPL (CC, f), SEOR1 (SE, g), CALS7 (SE, h), and DWF4 (GSK3-induced, i) in VISUAL-CC samples cultured with various bikinin concentrations for 4 days. Dots indicate relative expression levels for each sample. Statistical differences between samples are indicated by different letters (ANOVA, Tukey–Kramer method, n = 6). j Positive correlation between expression of SUC2 (CC) and DWF4 (GSK3-induced). k Negative correlation between expression of SUC2 (CC) and TCH4 (GSK3-suppressed). l Schematic model showing dose-dependent regulation of the SE/CC ratio by GSK3 activity.
SE/CC cell fates mostly depend on GSK3 activity
To identify the determinants of SE/CC cell fate, we investigated the impact of auxin and bikinin, as their concentrations differed between VISUAL and VISUAL-CC media (Fig. 1c). Exogenous auxin reduced pSUC2:ELUC expression in a dose-dependent manner (Supplementary Fig. 7a). Although SUC2 expression levels showed a similar decreased trend (Supplementary Fig. 7b), SEOR1 expression was not affected by auxin level (Supplementary Fig. 7c). Similarly, bikinin showed a repressive effect on pSUC2:ELUC expression levels (Fig. 3d). Unlike auxin, treatment with bikinin decreased expression of SUC2 in a dose-dependent manner while simultaneously increasing SEOR1 expression (Fig. 3e–h). Furthermore, two other CC and SE markers, SAPL and CALLOSE SYNTHASE 7 (CALS7)18 showed a similar response against bikinin treatment (Fig. 3e–h). Bikinin is a specific inhibitor of plant GSK3s19, and thus these results suggest that GSK3 activity plays a role in determining the SE/CC ratio. GSK3 activity correlates with the expression of BR biosynthetic genes, as a negative feedback regulation. Indeed, DWARF4 (DWF4)20, one of BR biosynthetic genes, was downregulated following bikinin treatment in a dose-dependent manner (Fig. 3i). In the VISUAL-CC transcriptome, expression of BR biosynthetic genes tended to be higher in the S samples and lower in the M samples (Supplementary Fig. 8), also suggesting the relationship between the GSK3 activity and the SE/CC ratio. Previous studies have reported 113 bikinin-suppressed genes19; then we investigated the correlation between these genes and VC genes in VISUAL-CC transcriptome data. The bikinin-suppressed genes showed higher expression in the S samples (Fig. 2j) and their expression exhibited positive correlation with expression of VC genes (Supplementary Fig. 5c; r = 0.94, P < 0.01). To investigate the relationship further, we quantitatively compared the expression patterns of SUC2 and GSK3-affected genes in the S and M samples. Expression of DWF4, a typical GSK3-induced gene (Fig. 3i), showed a strong positive correlation with SUC2 expression (Fig. 3j, r = 0.89, P < 0.005). Similarly, other GSK3-induced genes such as CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD) and BRASSINOSTEROID-6-OXIDASE 2 (BR6ox2) showed significantly higher expression in the S samples than in the M samples21,22 (Supplementary Fig. 6b). By contrast, expression of TOUCH 4 (TCH4), a typical GSK3-suppressed gene23, showed a significant negative correlation with SUC2 expression (Fig. 3k, r = −0.72, P < 0.05). These results suggest that the ratio of induced SE and -CC is largely dependent on GSK3 activity in VISUAL-CC (Fig. 3l).
Manipulation of GSK3 activity alters in vivo SE/CC ratio
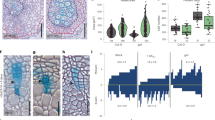
Then, we analyzed the role of GSK3s in in vivo secondary phloem development in Arabidopsis hypocotyls. In hypocotyls, SEs are characterized by vacant cytoplasm, whereas CCs are deeply stained with toluidine blue and they usually appear as pairs in a transverse section (Fig. 4a). Inhibition of GSK3 activity by bikinin treatment induced clusters of SEs and far fewer CCs (Fig. 4b). Bikinin treatment consistently reduced expression of pSUC2:YFPnls and resulted in clusters of pSEOR1:SEOR1-RFP signals in the dual phloem marker line (Fig. 4c, d), indicating that bikinin promotes SE formation and decreased CC number in vivo. Next, we confirmed the function of GSK3 proteins genetically using knockout mutants of members of the SKII subfamily (BIN2, BIL1, and BIL2) and RNAi knockdown for SKI subfamily members (AtSK11, AtSK12, and AtSK13)24, because they are the main targets of bikinin17. The phloem tissue of the bin2 bil1 bil2 AtSK13RNAi quadruple mutant exhibited a slight but significant decrease in CC occupancy (40%) when compared with wild-type plants (44%) (Fig. 4e, f, h). The gsk hextuple mutant (quadruple + AtSK11, AtSK12RNAi) showed a reduction in CC occupancy (20%), resulting in more SEs and few CCs (Fig. 4g, h). Moreover, in the hextuple mutant, some of the PPP cells unexpectedly differentiated into ectopic SE-like cells (Fig. 4g). Previous studies have revealed that the vascular cells express SKII subgroup genes BIN2 and BIN2-LIKE2 (BIL2)24. In addition, expression of SKI subgroup genes pSK11:GUS and pSK12:GUS25 was found in the vasculature, including the phloem tissue (Fig. 4j, k). Similarly to the GUS expression analysis, SKI/II gene expression was kept high in VISUAL time-course and in VISUAL-CC transcriptome data (Supplementary Fig. 9), indicating that six GSK3 members are present during phloem development. Next, we investigated local GSK3 activity in the vasculature using pDWF4:GUS, which is an indicator of high GSK3 activity. Supporting with our idea, pDWF4:GUS expression was detected in the phloem CCs but not in SEs (Fig. 4l). Taken together, our results indicate that GSK3 activity is required for maintaining high CC occupancy in planta.
a, b Toluidine blue-stained transverse sections through mock- (DMSO, a) and bikinin-treated (b) hypocotyls. SE, white empty cell; CC, dense purple cell. c, d Expression of pSEOR1:SEOR1-RFP (magenta) and pSUC2:YFPnls (green) in 10-day-old hypocotyls treated with DMSO (c) or 10 µM bikinin (d) for 10 days (n = 8). Confocal images of transverse (left) and longitudinal sections (right). e–g Toluidine blue-stained transverse sections for 10-day-old hypocotyls of Ws (wild type, e and gsk3 high-order mutant plants (gsk3 quadruple, f and gsk3 hextuple, g). Arrowheads: ectopic SEs at the PPP position; the ratio of individuals showing ectopic SEs is marked on the upper right of the image. h, i Box-and-whisker plots of SE/CC ratios (%) in Ws and gsk3 high-order mutants (h), and WT plants treated with or without 10 µM bikinin (i) calculated from toluidine blue-stained sections. Median values were indicated by central lines. The first (Q1) and third (Q3) quartile were shown as a box. Lines show the range of Q1 + 1.5× interquartile and Q3–1.5× interquartile. Dots indicated distributions of each sample. Numbers of individuals are marked (n = 12–26). Asterisks: significant differences determined using Dunnett’s or Student’s t test (**P < 0.005). j, k GUS staining of 10-day-old hypocotyls of pSK11:GUS (j) and pSK12:GUS (k) plants. l GUS staining of 11-day-old hypocotyls of pDWF4:GUS plants. GUS staining patterns on the cross section (left) and subsequent toluidine blue-stained section (right) were shown. GUS expression pattern was overlapped with toluidine blue staining. Scale bars: 10 µm (a–g); 20 µm (j, k); 50 µm (l).
GSK3s function as signaling hubs to control xylem differentiation in the cambium24. Here to focus on phloem development, bin2-1, a stable form of GSK326, was driven under promoters specific to each stage of phloem development. As we had previously demonstrated that the sequential genetic cascade in phloem SE differentiation is NAC020 (early), APL (middle), SEOR1 (late)2, we induced expression of bin2-1 under these different phloem promoters and investigated their phloem phenotype (Fig. 5a–f). Expression of bin2-1 driven by the APL and SEOR1 promoters did not affect phloem phenotypes, but pNAC020:bin2-1 slightly increased the ratio of CCs in the phloem (Fig. 5a–f). To objectively confirm the results with the CC marker, the number of pSUC2:YFPnls signal in WT and pNAC020:bin2-1 was quantified using a confocal microscope (Fig. 5g). YFP-positive cell number estimated from 3D-reconstruction images was significantly higher in the pNAC020:bin2-1 than in the WT (Fig. 5h–j), indicating that activation of GSK3 in the early phloem leads to increment of CC cell number. Taken together, our results suggest that GSK3s appear to function as cell-fate switches for determining differentiation into phloem CCs or SEs, and that GSK3 activity will be important for ensuring the proper ratio between CCs and SEs (Fig. 5k).
a–d Toluidine blue-stained transverse sections for hypocotyls of 11-day-old Col (wild type, a), pNAC020:bin2-1 (b), pAPL:bin2-1 (c), and pSEOR1:bin2-1 (d) plants. e Genetic cascade in phloem development revealed by VISUAL. f Box-and-whisker plots of SE/CC ratios (%) in wild type (WT), pNAC020:bin2-1, pAPL:bin2-1, and pSEOR1:bin2-1 calculated from toluidine blue-stained sections (n = 14–16). g Schematic illustration for 3D confocal imaging of pSUC2:YFPnls marker expression in hypocotyls. h, i Maximum intensity projection of pSUC2:YFPnls marker expression in the WT (h) and pNAC020:bin2-1 (i) was shown from XY (upper) and XZ (lower) angles. j Quantification of pSUC2:YFPnls-positive cells from 3D-reconstruction images in the WT and pNAC020:bin2-1 (n = 11–12). Asterisks: significant differences determined using Student’s t test (**P < 0.005). k Schematic showing that GSK3 activity determines the SE/CC ratio. For box-and-whisker plots, median values were indicated by central lines. The first (Q1) and third (Q3) quartile were shown as a box. Lines show the range of Q1 + 1.5× interquartile and Q3 − 1.5× interquartile. Dots indicated distributions of each sample. Scale bars: 10 µm (a–d); 50 µm (h, i).
BR-BES1 signaling is not involved in SE/CC-fate regulation
Finally, we examined the involvement of BR in CC differentiation, because GSK3s function as signal mediators in BR signaling26,27. However, application of brassinolide (BL), an active BR, did not alter the SE/CC ratio in hypocotyls (Supplementary Fig. 10). Moreover, bes1 bzr1 loss-of-function and bes1-D bzr1-D gain-of-function mutants for BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAXOLE RESISTANT 1 (BZR1), which are well-known transcription factors phosphorylated by GSK3s in BR signaling28, exhibited a normal phloem development in terms of the SE/CC ratio (Supplementary Fig. 11). These results suggest the possibility that other signaling pathway(s) than BR participates in controlling the SE/CC ratio.
Discussion
Recent studies have revealed that CCs are not only of importance for phloem transport, but also act as a signal center integrating environmental information into the developmental program29. We have established VISUAL-CC as a powerful tool for analyzing the functional and developmental processes of CCs. During secondary vascular development, it has been widely believed that SEs and CCs are derived from the same phloem precursors via asymmetric cell division30. In VISUAL-CC, SEs and CCs were formed from vascular stem cells as neighboring complexes after several rounds of cell division, suggesting that VISUAL-CC may reflect the process of secondary phloem development. By taking advantages of VISUAL-CC, we identified GSK3s as key molecular switches to specify cell fate toward SE or CC. Indeed, gsk3 gain-of-function and loss-of-function mutants altered the ratio of SE/CC differentiation in the hypocotyl vasculature, and our expression analysis together with previous results consistently revealed that SKI/II subgroup members of GSK3s including BIN2 are expressed in the vascular tissues of hypocotyls24. As SEs lose their nuclei during the differentiation process, support from adjacent CCs is essential for their function. Thus, maintenance of the SE/CC ratio by GSK3s will be an important mechanism ensuring survival under various environmental conditions. Although GSK3s act as a central regulator of SE/CC development, genetic experiments suggested the involvement of other signaling than BR mediated by GSK3s in this regulation. GSK3s have been implicated in regulation of phloem development through the interaction with OCTOPUS (OPS), which genetically functions together with BREVIS RADIX (BRX)31. Further studies on such interacting proteins and reverse genetic approaches combined with VISUAL-CC transcriptome data will be helpful for elucidating a new signaling cascade controlling the SE/CC ratio.
GSK3s also function in animals as molecular switches determining differentiation into alternate cell types32, suggesting their common and important role as cell-fate switches. On the other hand, GSK3 activity regulates asymmetric cell division in the stomata lineage by interacting with polarly localized proteins, which are required for specifying stomatal cell fate33. Further investigation with the context of asymmetric cell division will uncover the extent to which GSK3s serve as a common mechanism determining cell fate.
Methods
Plant materials
Arabidopsis plants used in this study is Col-0 accession, except for gsk3 high-order mutants (Ws background). To construct the CC-reporter lines, approximately 2.0 kb of the SUC2 promoter region was cloned and then fused with ELUC (Toyobo) or YFP containing a nuclear localization signal. A genomic fragment of SEOR1, approximately 4.8-kb long and containing 1.6 kb of the promoter, was fused with RFP to make pSEOR1:SEOR1-RFP; this was subsequently transformed into pSUC2:YFPnls to generate the double phloem marker line. To construct pDWF4:GUS, approximately 1.9 kb of the DWF4 promoter region was cloned and introduced into pMDC163 vector to create a GUS-fusion gene.
Phloem-specific GSK3 activation lines were constructed by cloning bin2-1 (BIN2E263K) and fusing it with the NAC020 (2.4 kb), APL (2.9 kb), and SEOR1 (1.6 kb) promoters using the LR reaction (Thermo Fisher Scientific). The gsk quadruple and hextuple mutants (Ws accession) used in this study were as reported previously23. The smxl4 smxl5 mutants were as reported previously6,7. The bes1 bzr1 loss-of-function mutants and bes1-D bzr1-D gain-of-function mutants were as reported previously34. pSK11:GUS and pSK12:GUS lines were as reported previously25.
Luciferase measurement
In this study, ELUC with PEST domain (Toyobo) was used as a short half-life luminescent protein. pSUC2:ELUC seedlings were co-cultured with 200 µM d-luciferin (Wako) in white 24-well plates (PerkinElmer). The time course of luciferase (LUC) activity was measured automatically using a TriStar2 LB942 (Berthold) within a growth chamber (Nihonika).
Microscopic observation
For deep imaging with confocal microscopes, isolated tissue samples were fixed for 3 h under vacuum in a fixative solution (4% paraformaldehyde and 0.01% Triton X-100 in 1× phosphate-buffered saline (PBS)). Fixed samples were washed twice with 1× PBS and transferred to ClearSee solution (25% urea, 15% xylitol, and 10% sodium deoxycholate). ClearSee solution was replaced with fresh solution every 2 days for 3–4 weeks. Calcofluor staining was performed 1 week before microscopic observations by adding 0.1% (w/v) calcofluor white to the ClearSee solution. The samples were stained overnight and then washed with ClearSee solution without calcofluor. Once the samples were stained, washing was continued as described above. Cleared samples were observed using LSM880 (Zeiss) or FV1200 (Olympus) confocal microscopes with Z stack. For the quantification of YFP-positive cells, we counted the number of cells in a phloem pole of approximately 420-µm length of hypocotyls based on reconstructed 3D images.
Electron microscopy
Sample preparation for electron microscopy observation was modified slightly from a previous study35. Briefly, leaf disks induced by VISUAL-CC were fixed and embedded in resin. Thin sections (100 nm) were mounted on glass slides. Sections were stained with 0.4% uranyl acetate solution (UA) and a lead citrate solution (Pb), and then coated with osmium tetroxide. Observations of slides were made using a FE-SEM (Hitachi SU 8220). Thinner (80 nm) sections were mounted on formvar-coated 1-slot copper grids, stained with 4% UA and Pb, and then observed using an 80-kV transmission electron microscope (JEOL JEM-1400 Flash).
qRT-PCR and microarray experiments
Total RNA was extracted from four cotyledons using RNeasy plant mini kit (Qiagen) after LUC measurement. After reverse transcription reaction, qRT-PCR was performed using LightCycler 480II (Roche) by a universal probe method. The expression value was normalized with an internal control UBQ14. Microarray experiments were conducted with the Arabidopsis Gene 1.0 ST Array (Affymetrix) and analyzed with Subio platform and R gplots package. Primers used in this study are listed in Supplementary Table 2.
Cross-section
Hypocotyls of 10- or 11-day-old seedlings were fixed with FAA (formalin:acetic acid:alcohol, 1:1:18) for 1 day. Fixed hypocotyls were subjected to an ethanol series (50%, 70%, 80%, 90%, and 99.5%) each for 30 min and then transferred into Technovit 7100 solution without Hardener II for 1 day. After the preincubation, samples were embedded in a mixture of Technovit 7100 + Hardener II (12.5:1) and incubated at 37 °C for more than 1 h to harden. Technovit samples were sliced into 2-µm sections using a LEICA RM2255 microtome and stained with 0.1% toluidine blue to enable CCs (deep purple) to be distinguished from SEs (white) under microscopy. We counted the CC/SE cell number per phloem pole on the sections and statistically analyzed the difference using more than 12 samples. Cross sections for GUS-stained samples were made as reported previously24.
Statistics and reproducibility
Microarray analysis was performed using three biologically independent samples for S, M, and V. Correlation among the samples was analyzed by Pearson correlation coefficient. For the quantification of cross-section or confocal images, at least two biologically independent samples were used. Statistical differences were computed by Student’s t test or ANOVA (Tukey–Kramer method).
VISUAL-CC
This protocol was a modified one from the previous VISUAL method36. VISUAL-CC consists of two distinct steps: vascular stem cell formation and subsequent phloem differentiation. As the initial step, 6- or 7-day-old seedlings were cultured with the conventional VISUAL medium for 2 days in order to induce sufficient amount of (pro)cambial cells. After that, samples are transferred into VISUAL-CC medium for SE–CC complex differentiation.
Materials for growth of plant samples before VISUAL induction
-
1.
MS growth medium: It contains 2.2 g/L MS Basal Medium (Sigma), 10 g/L sucrose, and 0.5 g/L 2-morpholinoethanesulfonic acid monohydrate (MES) in Milli-Q water and the pH is adjusted to 5.7 with KOH. The solution is autoclaved at 120 °C for 20 min and can be stored at room temperature up to several weeks.
-
2.
Sterilizing solution: Sodium hypochlorite solution is diluted in Milli-Q water in the ratio 1:9 (v/v) and 0.1% of Triton X-100 is added. This solution is prepared immediately before the sterilizing procedure.
-
3.
Sterilized 6-well plate (Sumilon).
-
4.
Autoclaved Milli-Q water.
-
5.
Surgical tape.
-
6.
Continuous light chamber (22 °C, 45–55 µmol/m2/s).
-
7.
Rotary shaker (Taitec).
Materials for VISUAL and VISUAL-CC
-
1.
VISUAL base medium: It contains 2.2 g/L MS Basal Medium and 50 g/L D(+)-Glucose in Milli-Q water and the pH is adjusted to 5.7 with KOH. The solution is autoclaved at 120 °C for 20 min and can be stored at room temperature for several weeks.
-
2.
VISUAL-CC base medium: It contains 2.2 g/L MS Basal Medium and 10 g/L D(+)-Glucose in Milli-Q water and the pH is adjusted to 5.7 with KOH. The solution is autoclaved at 120 °C for 20 min and can be stored at room temperature for several weeks. Note that Glucose concentration is different from that of VISUAL base medium.
-
3.
2,4-D stock: About 2.5 g/L 2,4-D stock dissolved in autoclaved Milli-Q water and sterilized through 0.22-µm filter units. It is stored in small amounts in sampling tubes at −20 °C.
-
4.
Kinetin stock: About 0.5 g/L Kinetin stock dissolved in 0.1 M KOH and sterilized through 0.22-µm filter units. It is stored in small amounts in sampling tubes at −20 °C.
-
5.
Bikinin stock: About 10 mM Bikinin stock dissolved in DMSO and sterilized through 0.22-µm filter units. It is stored in small amounts in sampling tubes at −20 °C.
-
6.
Sterilized 12-well plate (Sumilon).
-
7.
Surgical forceps.
-
8.
Continuous light chamber (22 °C, 60–70 µmol/m2/s).
-
9.
Rotary shaker (Taitec).
Methods for growth of plant samples before VISUAL induction
-
1.
Sterilizing solution is added to the Arabidopsis seeds in 1.5-mL sampling tubes and gently mixed using a rotator for 5 min. The tubes are transferred inside a clean bench and allowed to stand for further 5 min. The sterilizing solution is then removed using a pipette, and the seeds are washed with autoclaved Milli-Q water three times. The seeds are soaked in water at 4 °C for 2 days to keep the germination timing constant.
-
2.
About 10 mL of the prepared MS growth medium is poured into each well of a 6-well plate. Seeds are sown at a density of 8–10 seeds/well containing the MS growth medium, and the plate is sealed with surgical tape. The well plate is incubated for 6–7 days under continuous light (22 °C, 45–55 µmol/m2/s) with shaking at 110 rpm on a rotary shaker.
Methods for VISUAL
-
1.
2,4-D stock, kinetin stock, and bikinin stock are defrosted at room temperature before use. The tubes are transferred inside a clean bench and added to the VISUAL base medium to obtain a final concentration of 1.25 mg/L 2,4-D, 0.25 mg/L kinetin, and 10 µM bikinin. About 2.5 mL of the above medium is then added into each well of a 12-well plate.
-
2.
A pair of sharp surgical forceps are used to cut the bottom half of Arabidopsis 6–7-day-old plants across the center of the hypocotyl and the roots are removed. About 4 of the Arabidopsis explants are then transferred carefully to each well containing the induction medium using forceps, and the 12-well plate is sealed with surgical tape. The explants are cultured for 2 days under continuous light (22 °C, 60–70 µmol/m2/s) with shaking at 110 rpm on a rotary shaker.
Methods for VISUAL-CC
-
1.
2,4-D stock, kinetin stock, and bikinin stock are defrosted at room temperature before use. The tubes are transferred inside a clean bench and added to the VISUAL-CC base medium to obtain a final concentration of 0.25 mg/L 2,4-D, 0.25 mg/L kinetin, and 1 µM bikinin. About 2.5 mL of the above medium is then added into each well of a 12-well plate. Note that auxin and bikinin concentration is decreased when compared with the VISUAL.
-
2.
VISUAL-induced samples were transferred into the new CC medium and then cultured for 4 days under dark conditions (22 °C) with shaking at 110 rpm on a rotary shaker. Note that light severely affects the CC differentiation ratio.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Accession number of microarray data for VISUAL-CC is GSE141037. The Supplementary information includes the 67 VC genes list characterized in this study as Supplementary Table 1.
References
Heo, J. O., Blob, B. & Helariutta, Y. Differentiation of conductive cells: a matter of life and death. Curr. Opin. Plant Biol. 35, 23–29 (2017).
Kondo, Y. et al. Vascular cell induction culture system using arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell 28, 1250–1262 (2016).
Stadler, R. & Sauer, N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109, 299–306 (1996).
Kurihara, D., Mizuta, Y., Sato, Y. & Higashiyama, T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015).
Froelich, D. R. et al. Phloem ultrastructure and pressure flow: sieve-element-occlusion-related agglomerations do not affect translocation. Plant Cell 23, 4428–4445 (2011).
Wallner, E. S. et al. Strigolactone- and Karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27, 1241–1247 (2017).
Wu, Y. Y. et al. DCL2- and RDR6-dependent transitive silencing of SMXL4 and SMXL5 in Arabidopsis dcl4 mutants causes defective phloem transport and carbohydrate over-accumulation. Plant J. 90, 1064–1078 (2017).
Miyashima, S. et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565, 490–494 (2019).
Zhang, C., Barthelson, R. A., Lambert, G. M. & Galbraith, D. W. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147, 30–40 (2008).
Zhu, Y., Liu, L., Shen, L. & Yu, H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2, 16075 (2016).
Tabata, R. et al. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 (2014).
Roberts, I. et al. CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J. Exp. Bot. 67, 4889–4899 (2016).
Zhao, C., Craig, J. C., Petzold, H. E., Dickerman, A. W. & Beers, E. P. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818 (2005).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Ross-Elliott, T. J. et al. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. Elife 6, e24125 (2017).
Turner, S. R. & Somerville, C. R. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701 (1997).
Bonke, M., Thitamadee, M. S., Mähönen, A. P., Hauser, M. T. & Helariutta, Y. APL regulates vascular tissue identity in Arabidopsis. Nature 426, 181–186 (2003).
Xie, B., Wang, X., Zhu, M., Zhang, Z. & Hong, Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 65, 1–14 (2011).
De Rybel, B. et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 26, 594–604 (2009).
Choe, S. et al. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243 (1998).
Szekeres, M. et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 (1996).
Kim, T. W. et al. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17, 2397–2412 (2005).
Xu, W. et al. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7, 1555–1567 (1995).
Kondo, Y. et al. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5, 3504 (2014).
Li, C., Zhang, B., Chen, B., Ji, L. & Yu, H. Site-specific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 9, 571 (2018).
Li, J., Nam, K. H., Vafeados, D. & Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22 (2001).
Li, J. & Nam, K. H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301 (2002).
Wang, Z. Y., Bai, M. Y., Oh, E. & Zhu, J. Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724 (2012).
Ohkubo, Y., Tanaka, M., Tabata, R., Ogawa-Ohnishi, M. & Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3, 17029 (2017).
Esau, K. The Phloem. Handbuch der Pflanzenanatomie, Vol. 5, no. 2 (eds W. Zimmermann, P. Ozenda, and H.D. Wulff), (Gebruder Borntraeger, Berlin, 1969).
Breda, A. S., Hazak, O. & Hardtke, C. S. Phosphosite charge rather than shootward localization determines OCTOPUS activity in root protophloem. Proc. Natl Acad. Sci. USA 114, E5721–E5730 (2017).
Kim, L. & Kimmel, A. R. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr. Opin. Genet. Dev. 10, 508–514 (2000).
Houbaert, A. et al. POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574–578 (2018).
Saito, M., Kondo, Y. & Fukuda, H. BES1 and BZR1 redundantly promote phloem and xylem differentiation. Plant Cell Physiol. 59, 590–600 (2018).
Sugiyama, Y. et al. A Rho-actin signaling pathway shapes cell wall boundaries in Arabidopsis xylem vessels. Nat. Commun. 10, 468 (2019).
Nurani, A. M., Kondo, Y. & Fukuda, H. Ectopic vascular induction in arabidopsis cotyledons for sequential analysis of phloem differentiation. Methods Mol. Biol. 1830, 149–159 (2018).
Acknowledgements
We thank Yuki Fukaya, Akiho Suizu, and Yukiko Sugisawa for technical assistance, and Enrico Scarpella, Thomas Greb, Ho-Ming Chen, and Hao Yu for sharing materials. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (17H06476 to Y.K. and 15H05958 to H.F.), and from the Japan Society for the Promotion of Science (17H05008 to Y.K. and 16H06377 to H.F.). Y.H. laboratory was funded by the Gatsby Foundation [GAT3395/PR3)]; the National Science Foundation BBSRC grant [BB/N013158/1]; University of Helsinki [award 799992091], the ERC Advanced Investigator Grant SYMDEV [No. 323052] and by Finnish Centre of Excellence in Molecular Biology of Primary Producers (Academy of Finland CoE program 2014-2019) decision #271832.
Author information
Authors and Affiliations
Contributions
Y.K. designed the experiment. T.T., S.O., M.N., Y.O., T.F., Masato S., Mayuko S., M.W., K.T., and Y.K. performed the experiments. H.F. and Y.H. shared materials and information. T.T., H.F., and Y.K. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tamaki, T., Oya, S., Naito, M. et al. VISUAL-CC system uncovers the role of GSK3 as an orchestrator of vascular cell type ratio in plants. Commun Biol 3, 184 (2020). https://doi.org/10.1038/s42003-020-0907-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-020-0907-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.